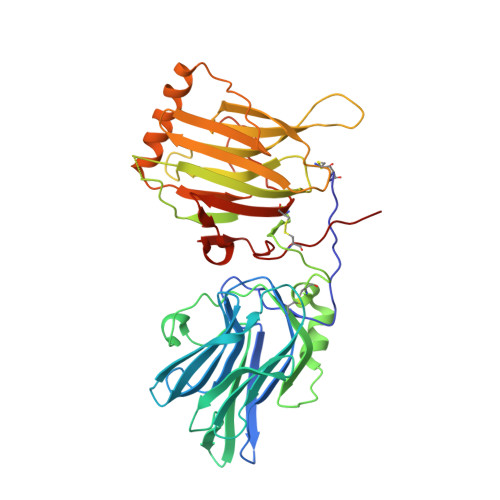
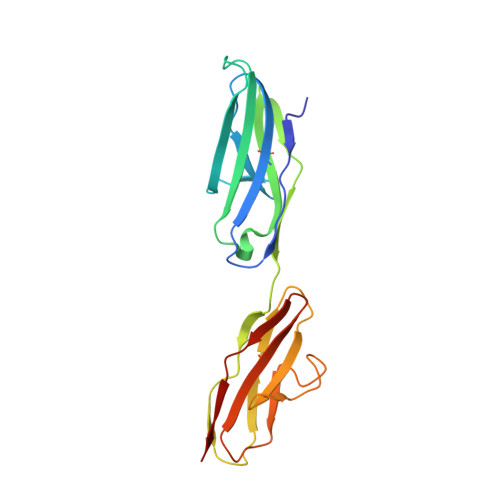
An engineered Axl 'decoy receptor' effectively silences the Gas6-Axl signaling axis.
Kariolis, M.S., Miao, Y.R., Jones, D.S., Kapur, S., Mathews, I.I., Giaccia, A.J., Cochran, J.R.(2014) Nat Chem Biol 10: 977-983
- PubMed: 25242553
- DOI: https://doi.org/10.1038/nchembio.1636
- Primary Citation of Related Structures:
4RA0 - PubMed Abstract:
Aberrant signaling through the Axl receptor tyrosine kinase has been associated with a myriad of human diseases, most notably metastatic cancer, identifying Axl and its ligand Gas6 as important therapeutic targets. Using rational and combinatorial approaches, we engineered an Axl 'decoy receptor' that binds Gas6 with high affinity and inhibits its function, offering an alternative approach from drug discovery efforts that directly target Axl. Four mutations within this high-affinity Axl variant caused structural alterations in side chains across the Gas6-Axl binding interface, stabilizing a conformational change on Gas6. When reformatted as an Fc fusion, the engineered decoy receptor bound Gas6 with femtomolar affinity, an 80-fold improvement compared to binding of the wild-type Axl receptor, allowing effective sequestration of Gas6 and specific abrogation of Axl signaling. Moreover, increased Gas6 binding affinity was critical and correlative with the ability of decoy receptors to potently inhibit metastasis and disease progression in vivo.
- Department of Bioengineering, Stanford University, Stanford, California, USA.
Organizational Affiliation: