Structural basis for RNA-genome recognition during bacteriophage Q beta replication.
Gytz, H., Mohr, D., Seweryn, P., Yoshimura, Y., Kutlubaeva, Z., Dolman, F., Chelchessa, B., Chetverin, A.B., Mulder, F.A., Brodersen, D.E., Knudsen, C.R.(2015) Nucleic Acids Res 43: 10893-10906
- PubMed: 26578560
- DOI: https://doi.org/10.1093/nar/gkv1212
- Primary Citation of Related Structures:
4R71 - PubMed Abstract:
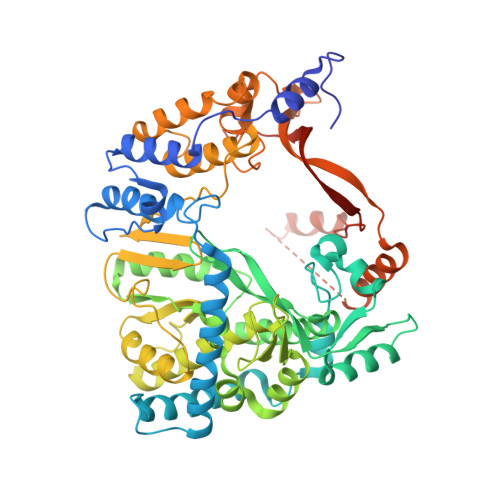
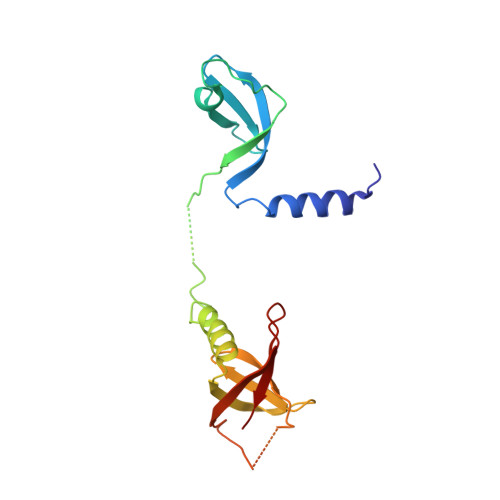
Upon infection of Escherichia coli by bacteriophage Qβ, the virus-encoded β-subunit recruits host translation elongation factors EF-Tu and EF-Ts and ribosomal protein S1 to form the Qβ replicase holoenzyme complex, which is responsible for amplifying the Qβ (+)-RNA genome. Here, we use X-ray crystallography, NMR spectroscopy, as well as sequence conservation, surface electrostatic potential and mutational analyses to decipher the roles of the β-subunit and the first two oligonucleotide-oligosaccharide-binding domains of S1 (OB1-2) in the recognition of Qβ (+)-RNA by the Qβ replicase complex. We show how three basic residues of the β subunit form a patch located adjacent to the OB2 domain, and use NMR spectroscopy to demonstrate for the first time that OB2 is able to interact with RNA. Neutralization of the basic residues by mutagenesis results in a loss of both the phage infectivity in vivo and the ability of Qβ replicase to amplify the genomic RNA in vitro. In contrast, replication of smaller replicable RNAs is not affected. Taken together, our data suggest that the β-subunit and protein S1 cooperatively bind the (+)-stranded Qβ genome during replication initiation and provide a foundation for understanding template discrimination during replication initiation.
- Department of Molecular Biology and Genetics, Aarhus University, DK-8000 Aarhus C, Denmark.
Organizational Affiliation: