The Crystal Structures of Middle East Respiratory Syndrome Coronavirus Papain-like Protease and Its Complex with an Inhibitor
Kong, L.Y., Wang, Q., Ming, Z.H., Sun, Y.N., Shaw, N., Yan, L.M., Lou, Z.Y., Rao, Z.H.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Non-structural protein 3 | A [auth B], B [auth A] | 336 | Betacoronavirus England 1 | Mutation(s): 0 Gene Names: 1a EC: 3.4.19.12 (PDB Primary Data), 3.4.22.69 (PDB Primary Data) | 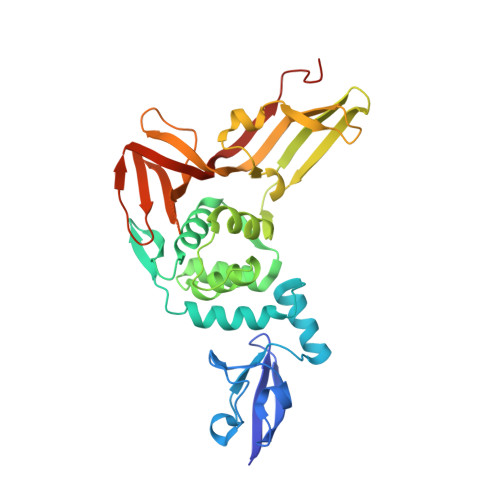 |
UniProt | |||||
Find proteins for K9N638 (Middle East respiratory syndrome-related coronavirus (isolate United Kingdom/H123990006/2012)) Explore K9N638 Go to UniProtKB: K9N638 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | K9N638 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| ZN Query on ZN | C [auth B], D [auth A] | ZINC ION Zn PTFCDOFLOPIGGS-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 136.624 | α = 90 |
| b = 136.624 | β = 90 |
| c = 239.518 | γ = 90 |
| Software Name | Purpose |
|---|---|
| HKL-2000 | data collection |
| PHENIX | model building |
| PHENIX | refinement |
| HKL-2000 | data reduction |
| HKL-2000 | data scaling |
| PHENIX | phasing |