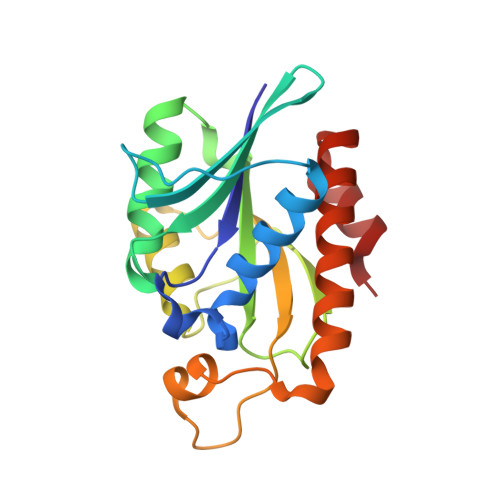
Crystal structure of peptidyl-tRNA hydrolase from a Gram-positive bacterium, Streptococcus pyogenes at 2.19 angstrom resolution shows the closed structure of the substrate-binding cleft.
Singh, A., Gautam, L., Sinha, M., Bhushan, A., Kaur, P., Sharma, S., Singh, T.P.(2014) FEBS Open Bio 4: 915-922
- PubMed: 25389518
- DOI: https://doi.org/10.1016/j.fob.2014.10.010
- Primary Citation of Related Structures:
4QT4 - PubMed Abstract:
Peptidyl-tRNA hydrolase (Pth) catalyses the release of tRNA and peptide components from peptidyl-tRNA molecules. Pth from a Gram-positive bacterium Streptococcus pyogenes (SpPth) was cloned, expressed, purified and crystallised. Three-dimensional structure of SpPth was determined by X-ray crystallography at 2.19 Å resolution. Structure determination showed that the asymmetric unit of the unit cell contained two crystallographically independent molecules, designated A and B. The superimposition of C(α) traces of molecules A and B showed an r.m.s. shift of 0.4 Å, indicating that the structures of two crystallographically independent molecules were identical. The polypeptide chain of SpPth adopted an overall α/β conformation. The substrate-binding cleft in SpPth is formed with three loops: the gate loop, Ile91-Leu102; the base loop, Gly108-Gly115; and the lid loop, Gly136-Gly150. Unlike in the structures of Pth from Gram-negative bacteria, the entry to the cleft in the structure of SpPth appeared to be virtually closed. However, the conformations of the active site residues were found to be similar.
- Department of Biophysics, All India Institute of Medical Sciences, New Delhi, India.
Organizational Affiliation: