Molecular imprint of exposure to naturally occurring genetic variants of human cytomegalovirus on the T cell repertoire.
Smith, C., Gras, S., Brennan, R.M., Bird, N.L., Valkenburg, S.A., Twist, K.A., Burrows, J.M., Miles, J.J., Chambers, D., Bell, S., Campbell, S., Kedzierska, K., Burrows, S.R., Rossjohn, J., Khanna, R.(2014) Sci Rep 4: 3993-3993
- PubMed: 24509977
- DOI: https://doi.org/10.1038/srep03993
- Primary Citation of Related Structures:
4QRS, 4QRT, 4QRU - PubMed Abstract:
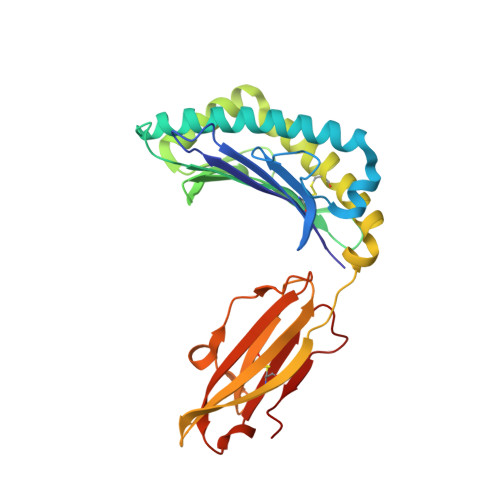
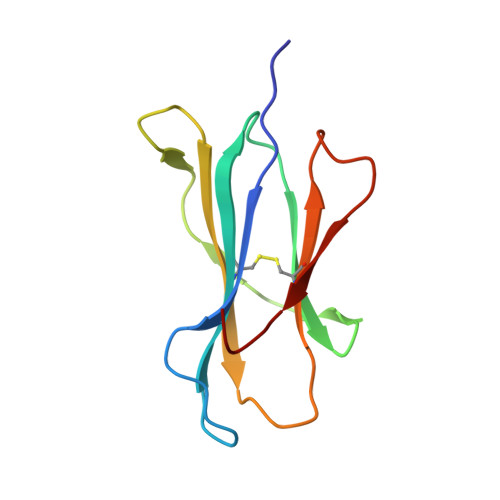
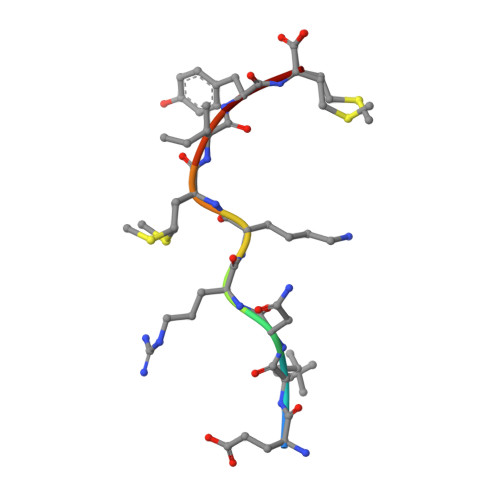
Exposure to naturally occurring variants of herpesviruses in clinical settings can have a dramatic impact on anti-viral immunity. Here we have evaluated the molecular imprint of variant peptide-MHC complexes on the T-cell repertoire during human cytomegalovirus (CMV) infection and demonstrate that primary co-infection with genetic variants of CMV was coincident with development of strain-specific T-cell immunity followed by emergence of cross-reactive virus-specific T-cells. Cross-reactive CMV-specific T cells exhibited a highly conserved public T cell repertoire, while T cells directed towards specific genetic variants displayed oligoclonal repertoires, unique to each individual. T cell recognition foot-print and pMHC-I structural analyses revealed that the cross-reactive T cells accommodate alterations in the pMHC complex with a broader foot-print focussing on the core of the peptide epitope. These findings provide novel molecular insight into how infection with naturally occurring genetic variants of persistent human herpesviruses imprints on the evolution of the anti-viral T-cell repertoire.
- QIMR Berghofer Medical Research Institute, Centre for Immunotherapy and Vaccine Development, Brisbane 4029 QLD Australia.
Organizational Affiliation: