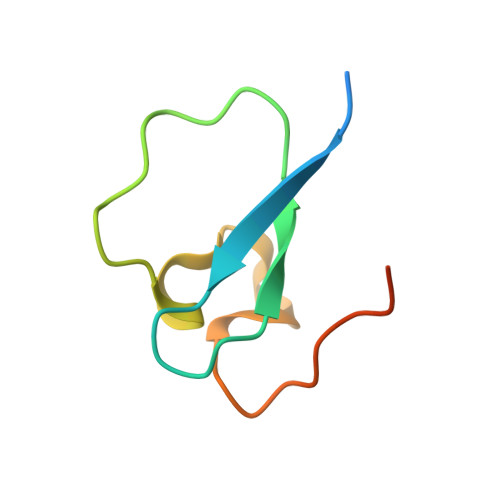
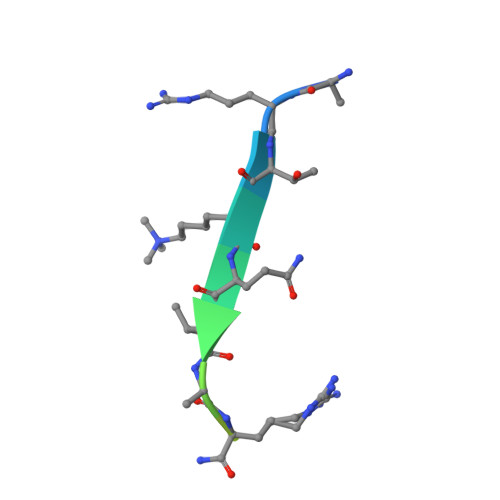
Family-wide Characterization of Histone Binding Abilities of Human CW Domain-containing Proteins.
Liu, Y., Tempel, W., Zhang, Q., Liang, X., Loppnau, P., Qin, S., Min, J.(2016) J Biological Chem 291: 9000-9013
- PubMed: 26933034
- DOI: https://doi.org/10.1074/jbc.M116.718973
- Primary Citation of Related Structures:
4O62, 4QQ4 - PubMed Abstract:
Covalent modifications of histone N-terminal tails play a critical role in regulating chromatin structure and controlling gene expression. These modifications are controlled by histone-modifying enzymes and read out by histone-binding proteins. Numerous proteins have been identified as histone modification readers. Here we report the family-wide characterization of histone binding abilities of human CW domain-containing proteins. We demonstrate that the CW domains in ZCWPW2 and MORC3/4 selectively recognize histone H3 trimethylated at Lys-4, similar to ZCWPW1 reported previously, while the MORC1/2 and LSD2 lack histone H3 Lys-4 binding ability. Our crystal structures of the CW domains of ZCWPW2 and MORC3 in complex with the histone H3 trimethylated at Lys-4 peptide reveal the molecular basis of this interaction. In each complex, two tryptophan residues in the CW domain form the "floor" and "right wall," respectively, of the methyllysine recognition cage. Our mutation results based on ZCWPW2 reveal that the right wall tryptophan residue is essential for binding, and the floor tryptophan residue enhances binding affinity. Our structural and mutational analysis highlights the conserved roles of the cage residues of CW domain across the histone methyllysine binders but also suggests why some CW domains lack histone binding ability.
- From the Structural Genomics Consortium, University of Toronto, Toronto, Ontario M5G 1L7, Canada, the Hubei Key Laboratory of Genetic Regulation and Integrative Biology, College of Life Science, Central China Normal University, Wuhan 430079, China.
Organizational Affiliation: