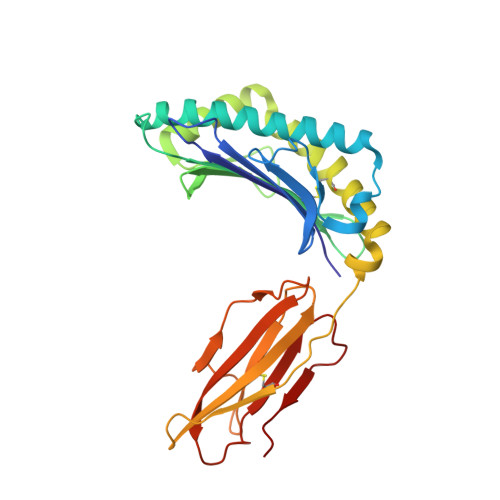
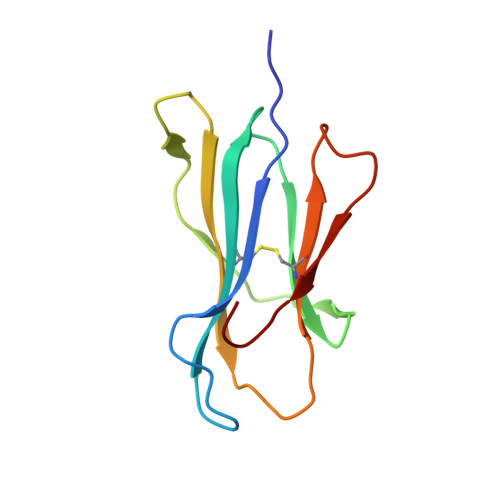
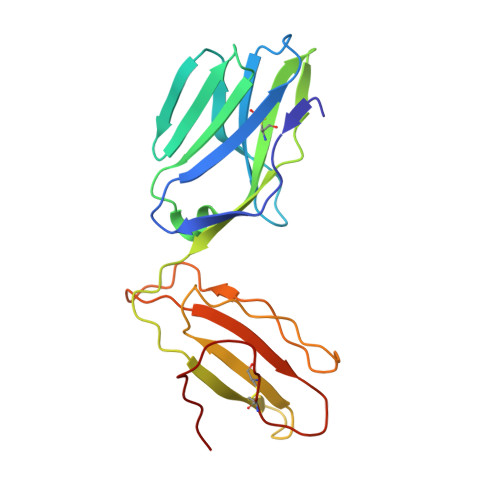
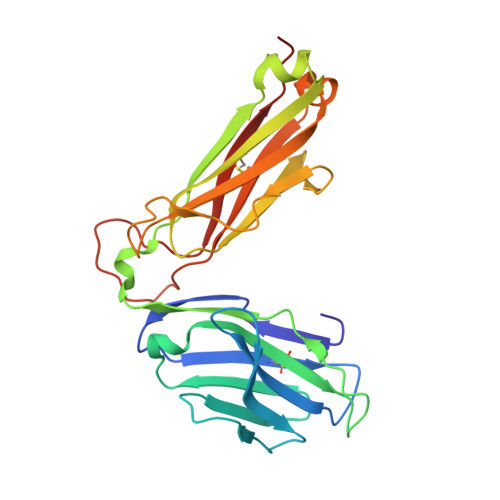
Structural basis for ineffective T-cell responses to MHC anchor residue-improved "heteroclitic" peptides.
Madura, F., Rizkallah, P.J., Holland, C.J., Fuller, A., Bulek, A., Godkin, A.J., Schauenburg, A.J., Cole, D.K., Sewell, A.K.(2015) Eur J Immunol 45: 584-591
- PubMed: 25471691
- DOI: https://doi.org/10.1002/eji.201445114
- Primary Citation of Related Structures:
4QOK - PubMed Abstract:
MHC anchor residue-modified "heteroclitic" peptides have been used in many cancer vaccine trials and often induce greater immune responses than the wild-type peptide. The best-studied system to date is the decamer MART-1/Melan-A26-35 peptide, EAAGIGILTV, where the natural alanine at position 2 has been modified to leucine to improve human leukocyte antigen (HLA)-A*0201 anchoring. The resulting ELAGIGILTV peptide has been used in many studies. We recently showed that T cells primed with the ELAGIGILTV peptide can fail to recognize the natural tumor-expressed peptide efficiently, thereby providing a potential molecular reason for why clinical trials of this peptide have been unsuccessful. Here, we solved the structure of a TCR in complex with HLA-A*0201-EAAGIGILTV peptide and compared it with its heteroclitic counterpart , HLA-A*0201-ELAGIGILTV. The data demonstrate that a suboptimal anchor residue at position 2 enables the TCR to "pull" the peptide away from the MHC binding groove, facilitating extra contacts with both the peptide and MHC surface. These data explain how a TCR can distinguish between two epitopes that differ by only a single MHC anchor residue and demonstrate how weak MHC anchoring can enable an induced-fit interaction with the TCR. Our findings constitute a novel demonstration of the extreme sensitivity of the TCR to minor alterations in peptide conformation.
- Division of Infection and Immunity, Cardiff University School of Medicine, Heath Park, Cardiff, UK.
Organizational Affiliation: