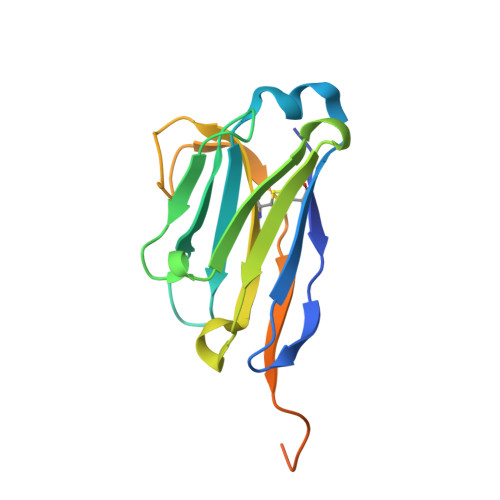
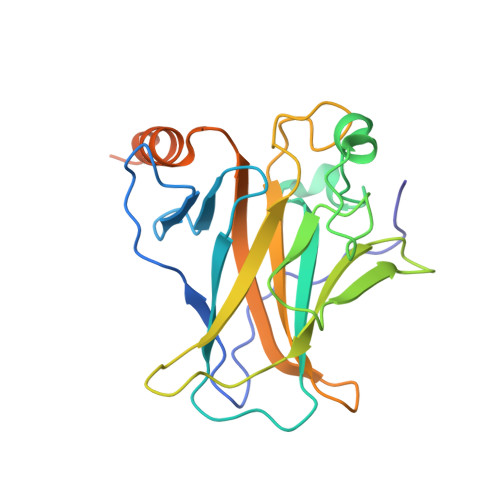
A nanobody modulates the p53 transcriptional program without perturbing its functional architecture.
Bethuyne, J., De Gieter, S., Zwaenepoel, O., Garcia-Pino, A., Durinck, K., Verhelle, A., Hassanzadeh-Ghassabeh, G., Speleman, F., Loris, R., Gettemans, J.(2014) Nucleic Acids Res 42: 12928-12938
- PubMed: 25324313
- DOI: https://doi.org/10.1093/nar/gku962
- Primary Citation of Related Structures:
4QO1 - PubMed Abstract:
The p53 transcription factor plays an important role in genome integrity. To perform this task, p53 regulates the transcription of genes promoting various cellular outcomes including cell cycle arrest, apoptosis or senescence. The precise regulation of this activity remains elusive as numerous mechanisms, e.g. posttranslational modifications of p53 and (non-)covalent p53 binding partners, influence the p53 transcriptional program. We developed a novel, non-invasive tool to manipulate endogenous p53. Nanobodies (Nb), raised against the DNA-binding domain of p53, allow us to distinctively target both wild type and mutant p53 with great specificity. Nb3 preferentially binds 'structural' mutant p53, i.e. R175H and R282W, while a second but distinct nanobody, Nb139, binds both mutant and wild type p53. The co-crystal structure of the p53 DNA-binding domain in complex with Nb139 (1.9 Å resolution) reveals that Nb139 binds opposite the DNA-binding surface. Furthermore, we demonstrate that Nb139 does not disturb the functional architecture of the p53 DNA-binding domain using conformation-specific p53 antibody immunoprecipitations, glutaraldehyde crosslinking assays and chromatin immunoprecipitation. Functionally, the binding of Nb139 to p53 allows us to perturb the transactivation of p53 target genes. We propose that reduced recruitment of transcriptional co-activators or modulation of selected post-transcriptional modifications account for these observations.
- Nanobody Lab, Department of Biochemistry, Ghent University, Albert Baertsoenkaai 3, B-9000 Ghent, Belgium.
Organizational Affiliation: