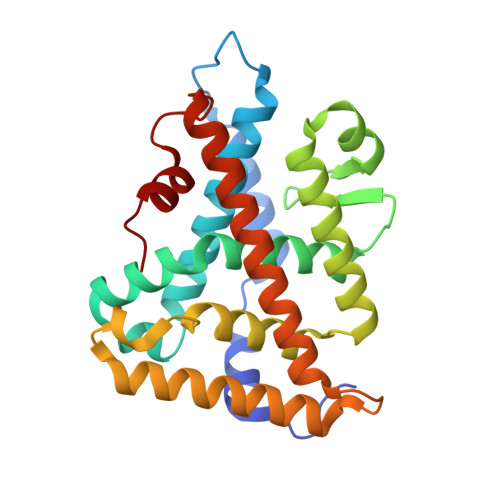
The signaling phospholipid PIP3 creates a new interaction surface on the nuclear receptor SF-1.
Blind, R.D., Sablin, E.P., Kuchenbecker, K.M., Chiu, H.J., Deacon, A.M., Das, D., Fletterick, R.J., Ingraham, H.A.(2014) Proc Natl Acad Sci U S A 111: 15054-15059
- PubMed: 25288771
- DOI: https://doi.org/10.1073/pnas.1416740111
- Primary Citation of Related Structures:
4QJR, 4QK4 - PubMed Abstract:
The signaling phosphatidylinositol lipids PI(4,5)P2 (PIP2) and PI(3,4,5)P3 (PIP3) bind nuclear receptor 5A family (NR5As), but their regulatory mechanisms remain unknown. Here, the crystal structures of human NR5A1 (steroidogenic factor-1, SF-1) ligand binding domain (LBD) bound to PIP2 and PIP3 show the lipid hydrophobic tails sequestered in the hormone pocket, as predicted. However, unlike classic nuclear receptor hormones, the phosphoinositide head groups are fully solvent-exposed and complete the LBD fold by organizing the receptor architecture at the hormone pocket entrance. The highest affinity phosphoinositide ligand PIP3 stabilizes the coactivator binding groove and increases coactivator peptide recruitment. This receptor-ligand topology defines a previously unidentified regulatory protein-lipid surface on SF-1 with the phosphoinositide head group at its nexus and poised to interact with other proteins. This surface on SF-1 coincides with the predicted binding site of the corepressor DAX-1 (dosage-sensitive sex reversal, adrenal hypoplasia critical region on chromosome X), and importantly harbors missense mutations associated with human endocrine disorders. Our data provide the structural basis for this poorly understood cluster of human SF-1 mutations and demonstrates how signaling phosphoinositides function as regulatory ligands for NR5As.
- Departments of Cellular and Molecular Pharmacology and.
Organizational Affiliation: