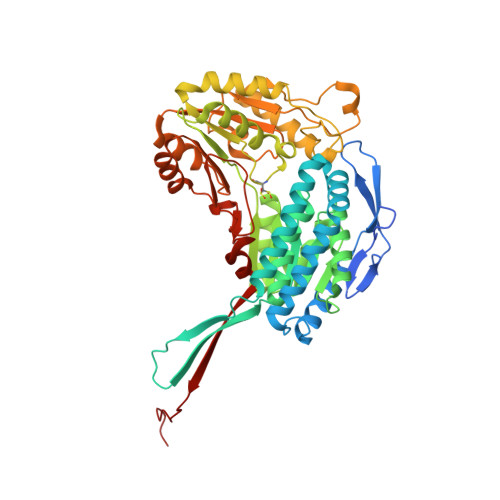
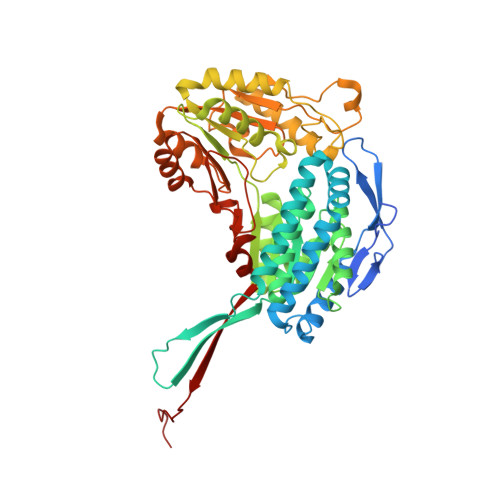
Structural and functional analysis of betaine aldehyde dehydrogenase from Staphylococcus aureus.
Halavaty, A.S., Rich, R.L., Chen, C., Joo, J.C., Minasov, G., Dubrovska, I., Winsor, J.R., Myszka, D.G., Duban, M., Shuvalova, L., Yakunin, A.F., Anderson, W.F.(2015) Acta Crystallogr D Biol Crystallogr 71: 1159-1175
- PubMed: 25945581
- DOI: https://doi.org/10.1107/S1399004715004228
- Primary Citation of Related Structures:
4NEA, 4NU9, 4Q92, 4QJE, 4QN2, 4QTO - PubMed Abstract:
When exposed to high osmolarity, methicillin-resistant Staphylococcus aureus (MRSA) restores its growth and establishes a new steady state by accumulating the osmoprotectant metabolite betaine. Effective osmoregulation has also been implicated in the acquirement of a profound antibiotic resistance by MRSA. Betaine can be obtained from the bacterial habitat or produced intracellularly from choline via the toxic betaine aldehyde (BA) employing the choline dehydrogenase and betaine aldehyde dehydrogenase (BADH) enzymes. Here, it is shown that the putative betaine aldehyde dehydrogenase SACOL2628 from the early MRSA isolate COL (SaBADH) utilizes betaine aldehyde as the primary substrate and nicotinamide adenine dinucleotide (NAD(+)) as the cofactor. Surface plasmon resonance experiments revealed that the affinity of NAD(+), NADH and BA for SaBADH is affected by temperature, pH and buffer composition. Five crystal structures of the wild type and three structures of the Gly234Ser mutant of SaBADH in the apo and holo forms provide details of the molecular mechanisms of activity and substrate specificity/inhibition of this enzyme.
- Department of Biochemistry and Molecular Genetics, Northwestern University, 303 East Chicago Avenue, Chicago, IL 60611, USA.
Organizational Affiliation: