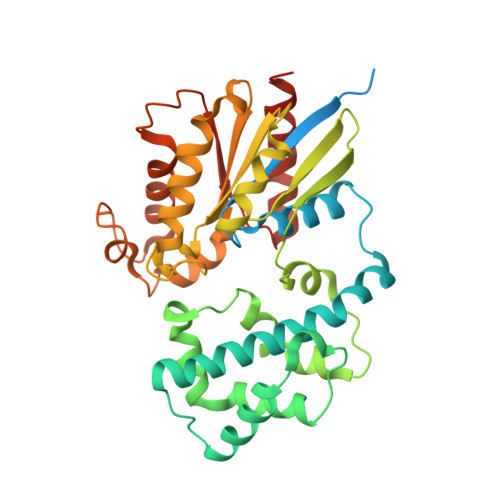
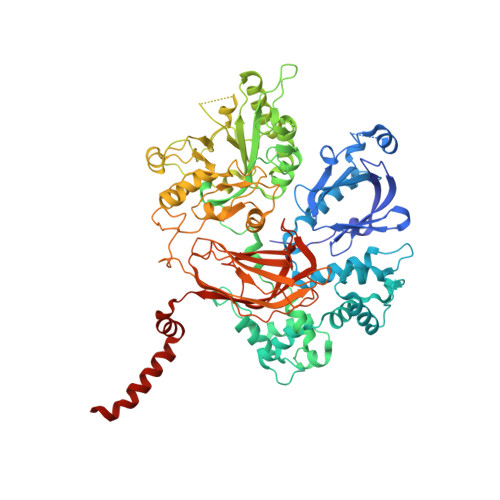
Molecular mechanisms of phospholipase C beta 3 autoinhibition.
Lyon, A.M., Begley, J.A., Manett, T.D., Tesmer, J.J.(2014) Structure 22: 1844-1854
- PubMed: 25435326
- DOI: https://doi.org/10.1016/j.str.2014.10.008
- Primary Citation of Related Structures:
4QJ3, 4QJ4, 4QJ5 - PubMed Abstract:
Phospholipase C β (PLCβ) enzymes are dramatically activated by heterotrimeric G proteins. Central to this response is the robust autoinhibition of PLCβ by the X-Y linker region within its catalytic core and by the Hα2' helix in the C-terminal extension of the enzyme. The molecular mechanism of each and their mutual dependence are poorly understood. Herein, it is shown that distinct regions within the X-Y linker have specific roles in regulating activity. Most important,an acidic stretch within the linker stabilizes a lid that occludes the active site, consistent with crystal structures of variants lacking this region. Inhibition by the Hα2' helix is independent of the X-Y linker and likely regulates activity by limiting membrane interaction of the catalytic core. Full activation of PLCβ thus requires multiple independent molecular events induced by membrane association of the catalytic core and by the binding of regulatory proteins.
- Life Sciences Institute, University of Michigan, 210 Washtenaw Avenue, Ann Arbor, MI 48109-2216, USA; Department of Pharmacology, University of Michigan, 1150 W. Medical Center Drive, 1301 MSRB III, Ann Arbor, MI 48109, USA; Department of Biological Chemistry, University of Michigan, 1150 W. Medical Center Drive, RM 5301 MSRB III, Ann Arbor, MI 48109-0600, USA.
Organizational Affiliation: