C2 domains as protein-protein interaction modules in the ciliary transition zone.
Remans, K., Burger, M., Vetter, I.R., Wittinghofer, A.(2014) Cell Rep 8: 1-9
- PubMed: 24981858
- DOI: https://doi.org/10.1016/j.celrep.2014.05.049
- Primary Citation of Related Structures:
4QAM - PubMed Abstract:
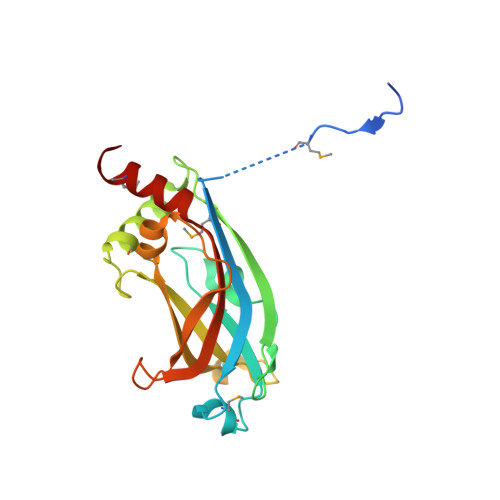
RPGR-interacting protein 1 (RPGRIP1) is mutated in the eye disease Leber congenital amaurosis (LCA) and its structural homolog, RPGRIP1-like (RPGRIP1L), is mutated in many different ciliopathies. Both are multidomain proteins that are predicted to interact with retinitis pigmentosa G-protein regulator (RPGR). RPGR is mutated in X-linked retinitis pigmentosa and is located in photoreceptors and primary cilia. We solved the crystal structure of the complex between the RPGR-interacting domain (RID) of RPGRIP1 and RPGR and demonstrate that RPGRIP1L binds to RPGR similarly. RPGRIP1 binding to RPGR affects the interaction with PDEδ, the cargo shuttling factor for prenylated ciliary proteins. RPGRIP1-RID is a C2 domain with a canonical β sandwich structure that does not bind Ca(2+) and/or phospholipids and thus constitutes a unique type of protein-protein interaction module. Judging from the large number of C2 domains in most of the ciliary transition zone proteins identified thus far, the structure presented here seems to constitute a cilia-specific module that is present in multiprotein transition zone complexes.
- Max Planck Institute of Molecular Physiology, Otto-Hahn-Strasse 11, 44227 Dortmund, Germany.
Organizational Affiliation: