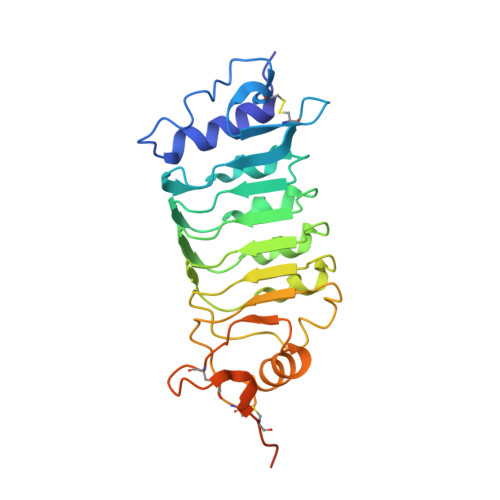
Structure of the OsSERK2 leucine-rich repeat extracellular domain.
McAndrew, R., Pruitt, R.N., Kamita, S.G., Pereira, J.H., Majumdar, D., Hammock, B.D., Adams, P.D., Ronald, P.C.(2014) Acta Crystallogr D Biol Crystallogr 70: 3080-3086
- PubMed: 25372696
- DOI: https://doi.org/10.1107/S1399004714021178
- Primary Citation of Related Structures:
4Q3G, 4Q3I - PubMed Abstract:
Somatic embryogenesis receptor kinases (SERKs) are leucine-rich repeat (LRR)-containing integral membrane receptors that are involved in the regulation of development and immune responses in plants. It has recently been shown that rice SERK2 (OsSERK2) is essential for XA21-mediated resistance to the pathogen Xanthomonas oryzae pv. oryzae. OsSERK2 is also required for the BRI1-mediated, FLS2-mediated and EFR-mediated responses to brassinosteroids, flagellin and elongation factor Tu (EF-Tu), respectively. Here, crystal structures of the LRR domains of OsSERK2 and a D128N OsSERK2 mutant, expressed as hagfish variable lymphocyte receptor (VLR) fusions, are reported. These structures suggest that the aspartate mutation does not generate any significant conformational change in the protein, but instead leads to an altered interaction with partner receptors.
- Physical Biosciences Division, Lawrence Berkeley National Laboratory, 1 Cyclotron Road, Berkeley, CA 94720, USA.
Organizational Affiliation: