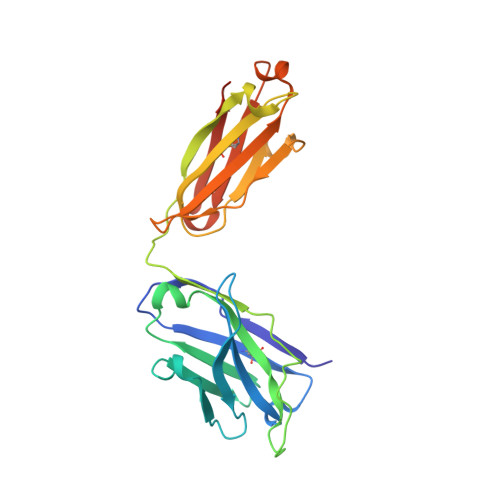
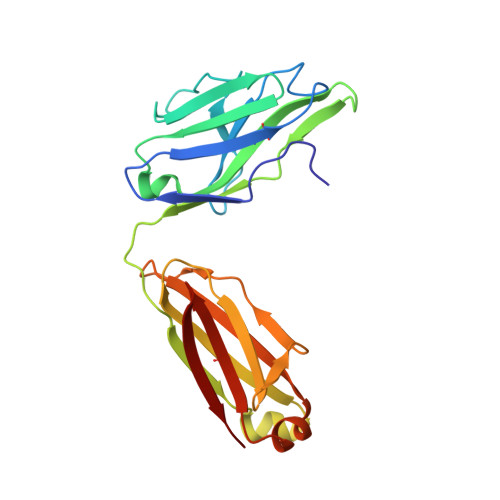
HIV-1 Receptor Binding Site-Directed Antibodies Using a VH1-2 Gene Segment Orthologue Are Activated by Env Trimer Immunization.
Navis, M., Tran, K., Bale, S., Phad, G.E., Guenaga, J., Wilson, R., Soldemo, M., McKee, K., Sundling, C., Mascola, J., Li, Y., Wyatt, R.T., Karlsson Hedestam, G.B.(2014) PLoS Pathog 10: e1004337-e1004337
- PubMed: 25166308
- DOI: https://doi.org/10.1371/journal.ppat.1004337
- Primary Citation of Related Structures:
4Q2Z - PubMed Abstract:
Broadly neutralizing antibodies (bNAbs) isolated from chronically HIV-1 infected individuals reveal important information regarding how antibodies target conserved determinants of the envelope glycoprotein (Env) spike such as the primary receptor CD4 binding site (CD4bs). Many CD4bs-directed bNAbs use the same heavy (H) chain variable (V) gene segment, VH1-2*02, suggesting that activation of B cells expressing this allele is linked to the generation of this type of Ab. Here, we identify the rhesus macaque VH1.23 gene segment to be the closest macaque orthologue to the human VH1-2 gene segment, with 92% homology to VH1-2*02. Of the three amino acids in the VH1-2*02 gene segment that define a motif for VRC01-like antibodies (W50, N58, flanking the HCDR2 region, and R71), the two identified macaque VH1.23 alleles described here encode two. We demonstrate that immunization with soluble Env trimers induced CD4bs-specific VH1.23-using Abs with restricted neutralization breadth. Through alanine scanning and structural studies of one such monoclonal Ab (MAb), GE356, we demonstrate that all three HCDRs are involved in neutralization. This contrasts to the highly potent CD4bs-directed VRC01 class of bNAb, which bind Env predominantly through the HCDR2. Also unlike VRC01, GE356 was minimally modified by somatic hypermutation, its light (L) chain CDRs were of average lengths and it displayed a binding footprint proximal to the trimer axis. These results illustrate that the Env trimer immunogen used here activates B cells encoding a VH1-2 gene segment orthologue, but that the resulting Abs interact distinctly differently with the HIV-1 Env spike compared to VRC01.
- Department of Microbiology, Tumor and Cell Biology, Karolinska Institutet, Stockholm, Sweden.
Organizational Affiliation: