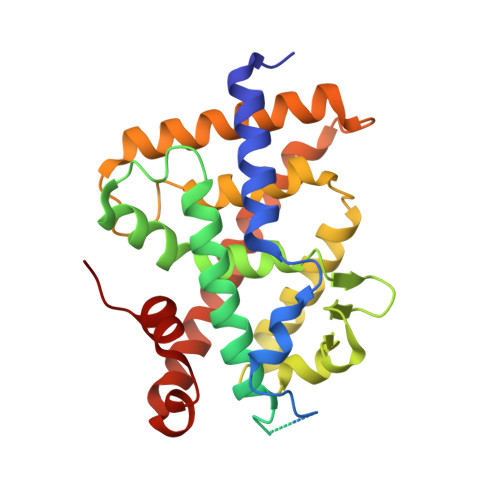
Structural insights into the molecular mechanism of vitamin d receptor activation by lithocholic Acid involving a new mode of ligand recognition.
Belorusova, A.Y., Eberhardt, J., Potier, N., Stote, R.H., Dejaegere, A., Rochel, N.(2014) J Med Chem 57: 4710-4719
- PubMed: 24818857
- DOI: https://doi.org/10.1021/jm5002524
- Primary Citation of Related Structures:
4Q0A - PubMed Abstract:
The vitamin D receptor (VDR), an endocrine nuclear receptor for 1α,25-dihydroxyvitamin D3, acts also as a bile acid sensor by binding lithocholic acid (LCA). The crystal structure of the zebrafish VDR ligand binding domain in complex with LCA and the SRC-2 coactivator peptide reveals the binding of two LCA molecules by VDR. One LCA binds to the canonical ligand-binding pocket, and the second one, which is not fully buried, is anchored to a site located on the VDR surface. Despite the low affinity of the alternative site, the binding of the second molecule promotes stabilization of the active receptor conformation. Biological activity assays, structural analysis, and molecular dynamics simulations indicate that the recognition of two ligand molecules is crucial for VDR agonism by LCA. The unique binding mode of LCA provides clues for the development of new chemical compounds that target alternative binding sites for therapeutic applications.
- Department of Integrative Structural Biology, Institut de Génétique et de Biologie Moléculaire et Cellulaire (IGBMC), Institut National de Santé et de Recherche Médicale (INSERM) U964, Centre National de Recherche Scientifique (CNRS) UMR 7104, Université de Strasbourg, 67404 Illkirch, France.
Organizational Affiliation: