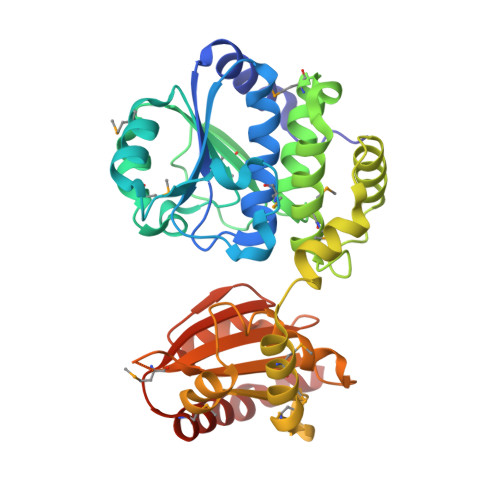
Crystal structure of an exopolyphosphatase-related protein from Bacteroides Fragilis. Northeast Structural Genomics target BFR192
Seetharaman, J., Abashidze, M., Wang, H., Janjua, H., Foote, E.L., Xiao, R., Nair, R., Everett, J.K., Acton, T.B., Rost, B., Montelione, G.T., Tong, L., Hunt, J.F., Northeast Structural Genomics Consortium (NESG)To be published.