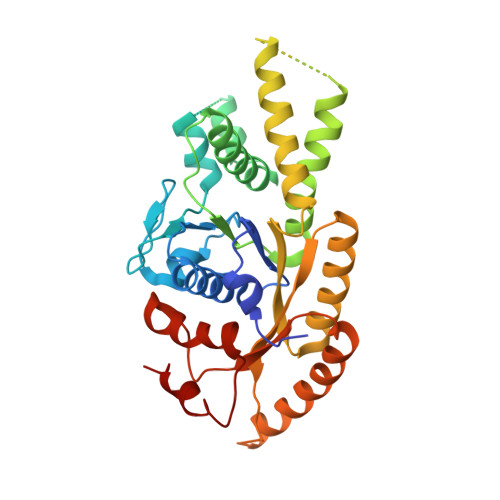
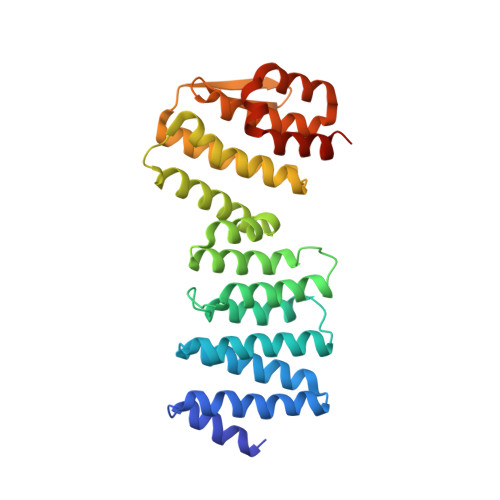
Crystal structure of ATP-bound Get3-Get4-Get5 complex reveals regulation of Get3 by Get4.
Gristick, H.B., Rao, M., Chartron, J.W., Rome, M.E., Shan, S.O., Clemons, W.M.(2014) Nat Struct Mol Biol 21: 437-442
- PubMed: 24727835
- DOI: https://doi.org/10.1038/nsmb.2813
- Primary Citation of Related Structures:
4PWX - PubMed Abstract:
Correct localization of membrane proteins is essential to all cells. Chaperone cascades coordinate the capture and handover of substrate proteins from the ribosomes to the target membranes, yet the mechanistic and structural details of these processes remain unclear. Here we investigate the conserved GET pathway, in which the Get4-Get5 complex mediates the handover of tail-anchor (TA) substrates from the cochaperone Sgt2 to the Get3 ATPase, the central targeting factor. We present a crystal structure of a yeast Get3-Get4-Get5 complex in an ATP-bound state and show how Get4 primes Get3 by promoting the optimal configuration for substrate capture. Structure-guided biochemical analyses demonstrate that Get4-mediated regulation of ATP hydrolysis by Get3 is essential to efficient TA-protein targeting. Analogous regulation of other chaperones or targeting factors could provide a general mechanism for ensuring effective substrate capture during protein biogenesis.
- Division of Chemistry and Chemical Engineering, California Institute of Technology, Pasadena, California, USA.
Organizational Affiliation: