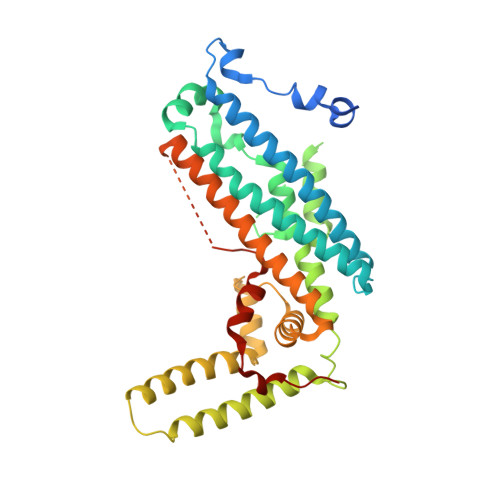
Structural and functional analyses of human tryptophan 2,3-dioxygenase
Meng, B., Wu, D., Gu, J., Ouyang, S., Ding, W., Liu, Z.J.(2014) Proteins 82: 3210-3216
- PubMed: 25066423
- DOI: https://doi.org/10.1002/prot.24653
- Primary Citation of Related Structures:
4PW8 - PubMed Abstract:
Tryptophan 2,3-dioxygenase (TDO), one of the two key enzymes in the kynurenine pathway, catalyzes the indole ring cleavage at the C2-C3 bond of L-tryptophan. This is a rate-limiting step in the regulation of tryptophan concentration in vivo, and is thus important in drug discovery for cancer and immune diseases. Here, we report the crystal structure of human TDO (hTDO) without the heme cofactor to 2.90 Å resolution. The overall fold and the tertiary assembly of hTDO into a tetramer, as well as the active site architecture, are well conserved and similar to the structures of known orthologues. Kinetic and mutational studies confirmed that eight residues play critical roles in L-tryptophan oxidation.
- National Laboratory of Biomacromolecules, Institute of Biophysics, Chinese Academy of Sciences, Beijing, 100101, China; University of Chinese Academy of Sciences, Beijing, 100049, China.
Organizational Affiliation: