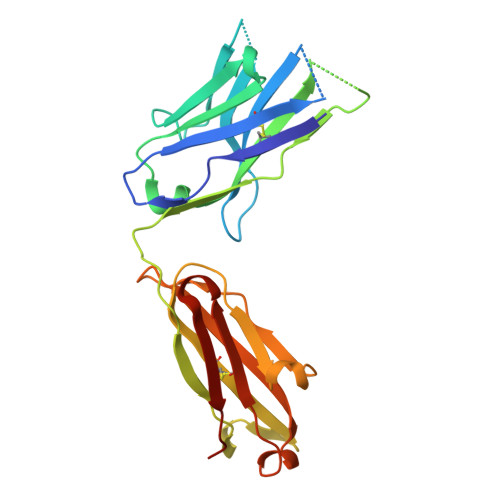
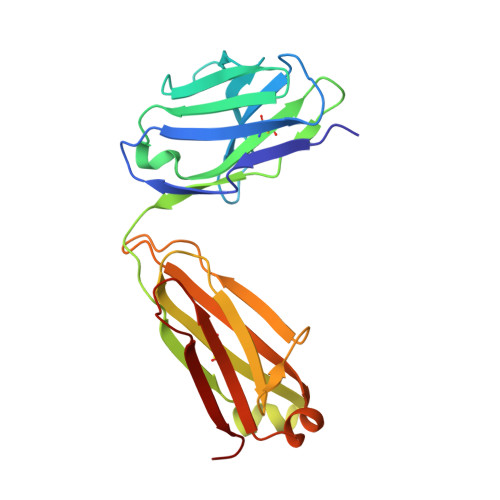
Inhibition of plasma kallikrein by a highly specific active site blocking antibody.
Kenniston, J.A., Faucette, R.R., Martik, D., Comeau, S.R., Lindberg, A.P., Kopacz, K.J., Conley, G.P., Chen, J., Viswanathan, M., Kastrapeli, N., Cosic, J., Mason, S., DiLeo, M., Abendroth, J., Kuzmic, P., Ladner, R.C., Edwards, T.E., TenHoor, C., Adelman, B.A., Nixon, A.E., Sexton, D.J.(2014) J Biological Chem 289: 23596-23608
- PubMed: 24970892
- DOI: https://doi.org/10.1074/jbc.M114.569061
- Primary Citation of Related Structures:
4OGX, 4OGY, 4PUB - PubMed Abstract:
Plasma kallikrein (pKal) proteolytically cleaves high molecular weight kininogen to generate the potent vasodilator and the pro-inflammatory peptide, bradykinin. pKal activity is tightly regulated in healthy individuals by the serpin C1-inhibitor, but individuals with hereditary angioedema (HAE) are deficient in C1-inhibitor and consequently exhibit excessive bradykinin generation that in turn causes debilitating and potentially fatal swelling attacks. To develop a potential therapeutic agent for HAE and other pKal-mediated disorders, we used phage display to discover a fully human IgG1 monoclonal antibody (DX-2930) against pKal. In vitro experiments demonstrated that DX-2930 potently inhibits active pKal (Ki = 0.120 ± 0.005 nM) but does not target either the zymogen (prekallikrein) or any other serine protease tested. These findings are supported by a 2.1-Å resolution crystal structure of pKal complexed to a DX-2930 Fab construct, which establishes that the pKal active site is fully occluded by the antibody. DX-2930 injected subcutaneously into cynomolgus monkeys exhibited a long half-life (t½ ∼ 12.5 days) and blocked high molecular weight kininogen proteolysis in activated plasma in a dose- and time-dependent manner. Furthermore, subcutaneous DX-2930 reduced carrageenan-induced paw edema in rats. A potent and long acting inhibitor of pKal activity could be an effective treatment option for pKal-mediated diseases, such as HAE.
- From the Dyax Corp., Burlington, Massachusetts 01803.
Organizational Affiliation: