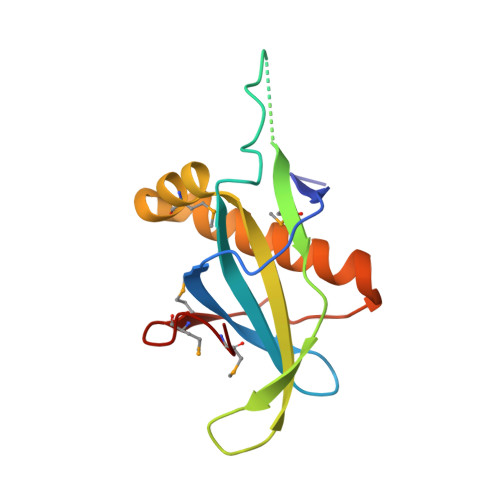
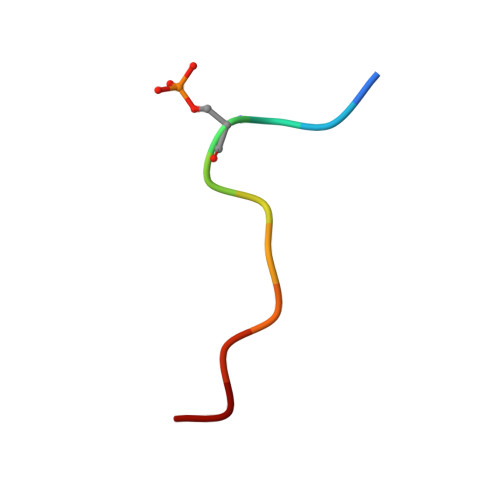
Phosphorylation-Dependent PIH1D1 Interactions Define Substrate Specificity of the R2TP Cochaperone Complex.
Horejsi, Z., Stach, L., Flower, T.G., Joshi, D., Flynn, H., Skehel, J.M., O'Reilly, N.J., Ogrodowicz, R.W., Smerdon, S.J., Boulton, S.J.(2014) Cell Rep 7: 19-26
- PubMed: 24656813
- DOI: https://doi.org/10.1016/j.celrep.2014.03.013
- Primary Citation of Related Structures:
4PSF, 4PSI - PubMed Abstract:
The R2TP cochaperone complex plays a critical role in the assembly of multisubunit machines, including small nucleolar ribonucleoproteins (snoRNPs), RNA polymerase II, and the mTORC1 and SMG1 kinase complexes, but the molecular basis of substrate recognition remains unclear. Here, we describe a phosphopeptide binding domain (PIH-N) in the PIH1D1 subunit of the R2TP complex that preferentially binds to highly acidic phosphorylated proteins. A cocrystal structure of a PIH-N domain/TEL2 phosphopeptide complex reveals a highly specific phosphopeptide recognition mechanism in which Lys57 and 64 in PIH1D1, along with a conserved DpSDD phosphopeptide motif within TEL2, are essential and sufficient for binding. Proteomic analysis of PIH1D1 interactors identified R2TP complex substrates that are recruited by the PIH-N domain in a sequence-specific and phosphorylation-dependent manner suggestive of a common mechanism of substrate recognition. We propose that protein complexes assembled by the R2TP complex are defined by phosphorylation of a specific motif and recognition by the PIH1D1 subunit.
- DNA Damage Response Laboratory, London Research Institute, Clare Hall, South Mimms EN6 3LD, UK.
Organizational Affiliation: