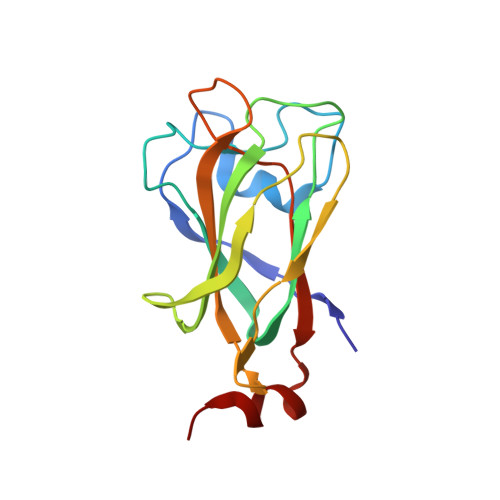
Structure of mouse muskelin discoidin domain and biochemical characterization of its self-association.
Kim, K.H., Hong, S.K., Hwang, K.Y., Kim, E.E.(2014) Acta Crystallogr D Biol Crystallogr 70: 2863-2874
- PubMed: 25372678
- DOI: https://doi.org/10.1107/S139900471401894X
- Primary Citation of Related Structures:
4PQQ - PubMed Abstract:
Muskelin is an intracellular kelch-repeat protein comprised of discoidin, LisH, CTLH and kelch-repeat domains. It is involved in cell adhesion and the regulation of cytoskeleton dynamics as well as being a component of a putative E3 ligase complex. Here, the first crystal structure of mouse muskelin discoidin domain (MK-DD) is reported at 1.55 Å resolution, which reveals a distorted eight-stranded β-barrel with two short α-helices at one end of the barrel. Interestingly, the N- and C-termini are not linked by the disulfide bonds found in other eukaryotic discoidin structures. A highly conserved MIND motif appears to be the determinant for MK-DD specific interaction together with the spike loops. Analysis of interdomain interaction shows that MK-DD binds the kelch-repeat domain directly and that this interaction depends on the presence of the LisH domain.
- Biomedical Research Institute, Korea Institute of Science and Technology, Hwarang-ro 14-gil 5, Seongbuk-gu, Seoul 136-791, Republic of Korea.
Organizational Affiliation: