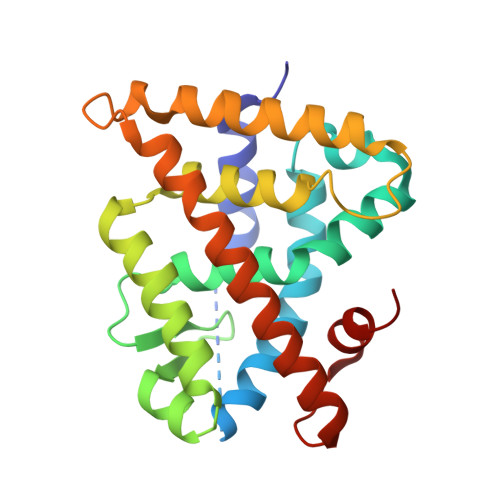
Methyl substitution of a rexinoid agonist improves potency and reveals site of lipid toxicity.
Atigadda, V.R., Xia, G., Desphande, A., Boerma, L.J., Lobo-Ruppert, S., Grubbs, C.J., Smith, C.D., Brouillette, W.J., Muccio, D.D.(2014) J Med Chem 57: 5370-5380
- PubMed: 24801499
- DOI: https://doi.org/10.1021/jm5004792
- Primary Citation of Related Structures:
4POH, 4POJ, 4PP3, 4PP5 - PubMed Abstract:
(2E,4E,6Z,8E)-8-(3',4'-Dihydro-1'(2'H)-naphthalen-1'-ylidene)-3,7-dimethyl-2,4,6-octatrienoic acid, 9cUAB30, is a selective rexinoid that displays substantial chemopreventive capacity with little toxicity. 4-Methyl-UAB30, an analogue of 9cUAB30, is a potent RXR agonist but caused increased lipid biosynthesis unlike 9cUAB30. To evaluate how methyl substitution influenced potency and lipid biosynthesis, we synthesized four 9cUAB30 homologues with methyl substitutions at the 5-, 6-, 7-, or 8-position of the tetralone ring. The syntheses and biological evaluations of these new analogues are reported here along with the X-ray crystal structures of each homologue bound to the ligand binding domain of hRXRα. We demonstrate that each homologue of 9cUAB30 is a more potent agonist, but only the 7-methyl-9cUAB30 caused severe hyperlipidemia in rats. On the basis of the X-ray crystal structures of these new rexinoids and bexarotene (Targretin) bound to hRXRα-LBD, we reveal that each rexinoid, which induced hyperlipidemia, had methyl groups that interacted with helix 7 residues of the LBD.
- Departments of †Chemistry, ‡Biochemistry and Molecular Genetics, §Medicine, and ∥Vision Sciences, University of Alabama at Birmingham , Birmingham, Alabama 35294, United States.
Organizational Affiliation: