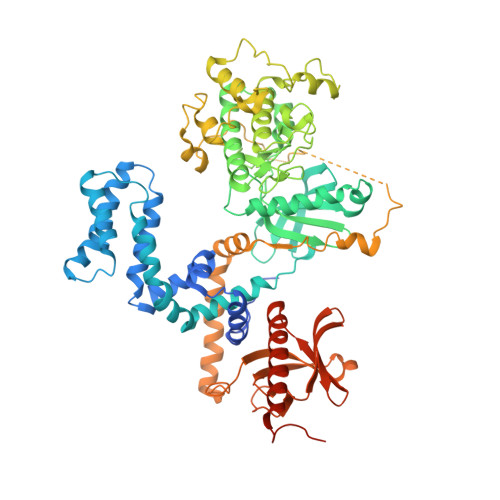
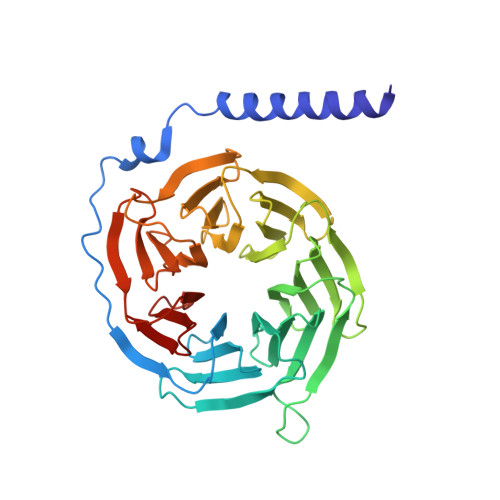
Identification and structure-function analysis of subfamily selective g protein-coupled receptor kinase inhibitors.
Homan, K.T., Larimore, K.M., Elkins, J.M., Szklarz, M., Knapp, S., Tesmer, J.J.(2015) ACS Chem Biol 10: 310-319
- PubMed: 25238254
- DOI: https://doi.org/10.1021/cb5006323
- Primary Citation of Related Structures:
4PNI, 4PNK - PubMed Abstract:
Selective inhibitors of individual subfamilies of G protein-coupled receptor kinases (GRKs) would serve as useful chemical probes as well as leads for therapeutic applications ranging from heart failure to Parkinson's disease. To identify such inhibitors, differential scanning fluorimetry was used to screen a collection of known protein kinase inhibitors that could increase the melting points of the two most ubiquitously expressed GRKs: GRK2 and GRK5. Enzymatic assays on 14 of the most stabilizing hits revealed that three exhibit nanomolar potency of inhibition for individual GRKs, some of which exhibiting orders of magnitude selectivity. Most of the identified compounds can be clustered into two chemical classes: indazole/dihydropyrimidine-containing compounds that are selective for GRK2 and pyrrolopyrimidine-containing compounds that potently inhibit GRK1 and GRK5 but with more modest selectivity. The two most potent inhibitors representing each class, GSK180736A and GSK2163632A, were cocrystallized with GRK2 and GRK1, and their atomic structures were determined to 2.6 and 1.85 Å spacings, respectively. GSK180736A, developed as a Rho-associated, coiled-coil-containing protein kinase inhibitor, binds to GRK2 in a manner analogous to that of paroxetine, whereas GSK2163632A, developed as an insulin-like growth factor 1 receptor inhibitor, occupies a novel region of the GRK active site cleft that could likely be exploited to achieve more selectivity. However, neither compound inhibits GRKs more potently than their initial targets. This data provides the foundation for future efforts to rationally design even more potent and selective GRK inhibitors.
- Life Sciences Institute and the Departments of Pharmacology and Biological Sciences, University of Michigan , Ann Arbor, Michigan 48109, United States.
Organizational Affiliation: