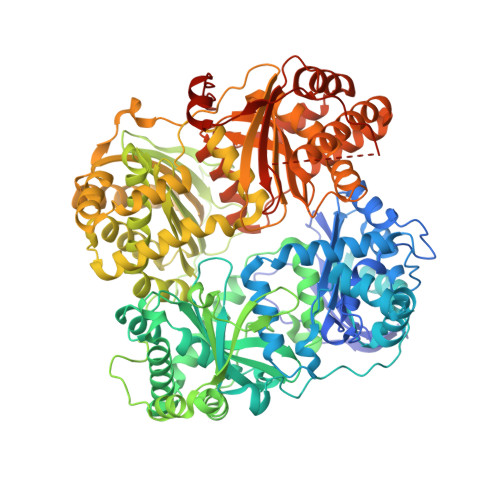
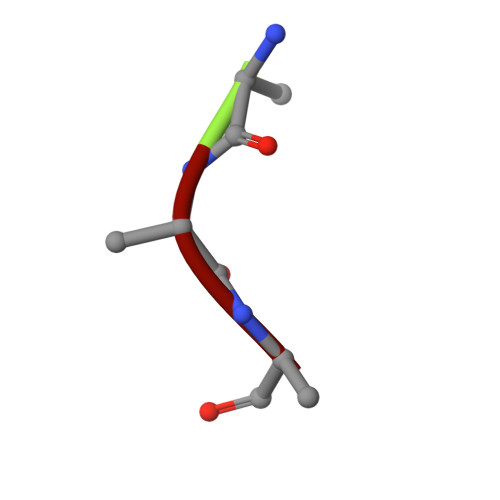
Crystal structure of IDE complexed with an inhibitor
Durham, T.B., Toth, J.L., Klimkowski, V.J., Cao, J.X.C., Siesky, A.M., Alexander-Chacko, J., Wu, G.Y., Dixon, J.T., McGee, J.E., Wang, Y., Guo, S., Cavitt, R.N., Schindler, J., Thibodeaux, S.J., Calvert, N.A., Coghlan, M.J., Sindelar, D.K., Christe, M., Kiselyov, V.V., Dodson, M., Sloop, K.W.To be published.