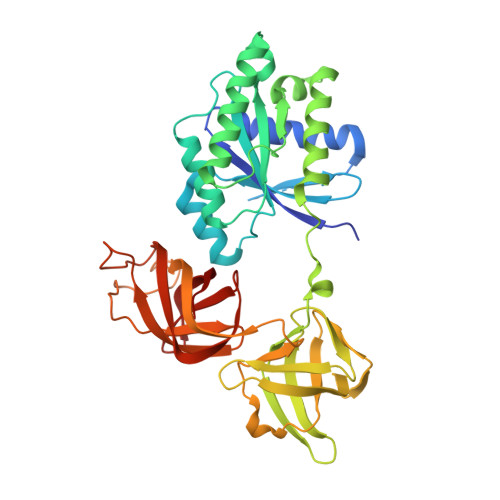
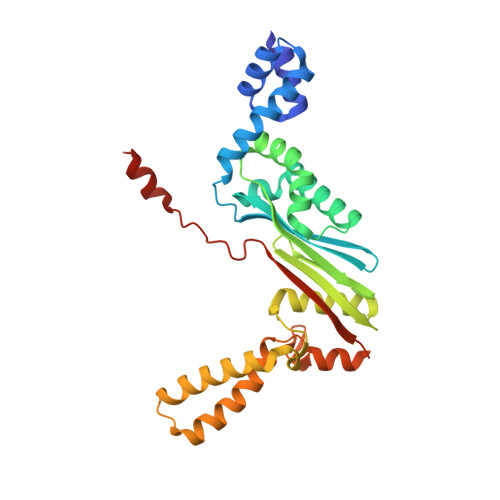
Structural outline of the detailed mechanism for elongation factor Ts-mediated guanine nucleotide exchange on elongation factor Tu.
Thirup, S.S., Van, L.B., Nielsen, T.K., Knudsen, C.R.(2015) J Struct Biol 191: 10-21
- PubMed: 26073967
- DOI: https://doi.org/10.1016/j.jsb.2015.06.011
- Primary Citation of Related Structures:
4PC1, 4PC2, 4PC3, 4PC6, 4PC7 - PubMed Abstract:
Translation elongation factor EF-Tu belongs to the superfamily of guanine-nucleotide binding proteins, which play key cellular roles as regulatory switches. All G-proteins require activation via exchange of GDP for GTP to carry out their respective tasks. Often, guanine-nucleotide exchange factors are essential to this process. During translation, EF-Tu:GTP transports aminoacylated tRNA to the ribosome. GTP is hydrolyzed during this process, and subsequent reactivation of EF-Tu is catalyzed by EF-Ts. The reaction path of guanine-nucleotide exchange is structurally poorly defined for EF-Tu and EF-Ts. We have determined the crystal structures of the following reaction intermediates: two structures of EF-Tu:GDP:EF-Ts (2.2 and 1.8Å resolution), EF-Tu:PO4:EF-Ts (1.9Å resolution), EF-Tu:GDPNP:EF-Ts (2.2Å resolution) and EF-Tu:GDPNP:pulvomycin:Mg(2+):EF-Ts (3.5Å resolution). These structures provide snapshots throughout the entire exchange reaction and suggest a mechanism for the release of EF-Tu in its GTP conformation. An inferred sequence of events during the exchange reaction is presented.
- Aarhus University, Department of Molecular Biology and Genetics, Center for Structural Biology, DK-8000 Aarhus C, Denmark. Electronic address: sth@mbg.au.dk.
Organizational Affiliation: