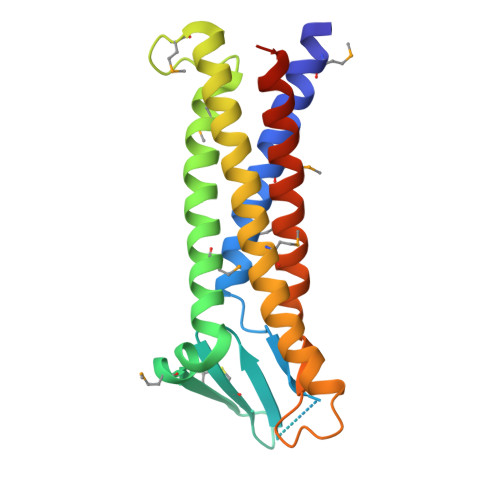
Crystal structure of a claudin provides insight into the architecture of tight junctions.
Suzuki, H., Nishizawa, T., Tani, K., Yamazaki, Y., Tamura, A., Ishitani, R., Dohmae, N., Tsukita, S., Nureki, O., Fujiyoshi, Y.(2014) Science 344: 304-307
- PubMed: 24744376
- DOI: https://doi.org/10.1126/science.1248571
- Primary Citation of Related Structures:
4P79 - PubMed Abstract:
Tight junctions are cell-cell adhesion structures in epithelial cell sheets that surround organ compartments in multicellular organisms and regulate the permeation of ions through the intercellular space. Claudins are the major constituents of tight junctions and form strands that mediate cell adhesion and function as paracellular barriers. We report the structure of mammalian claudin-15 at a resolution of 2.4 angstroms. The structure reveals a characteristic β-sheet fold comprising two extracellular segments, which is anchored to a transmembrane four-helix bundle by a consensus motif. Our analyses suggest potential paracellular pathways with distinctive charges on the extracellular surface, providing insight into the molecular basis of ion homeostasis across tight junctions.
- Cellular and Structural Physiology Institute, Nagoya University, Chikusa, Nagoya 464-8601, Japan.
Organizational Affiliation: