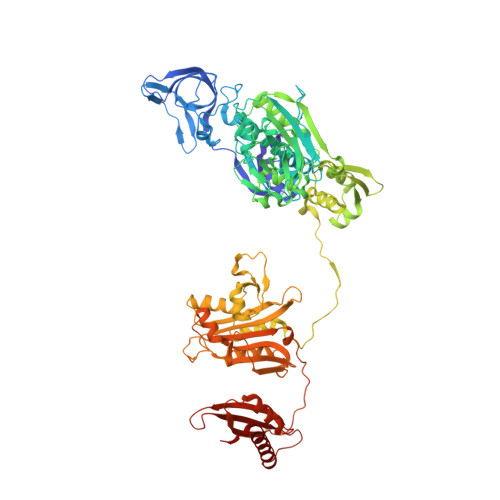
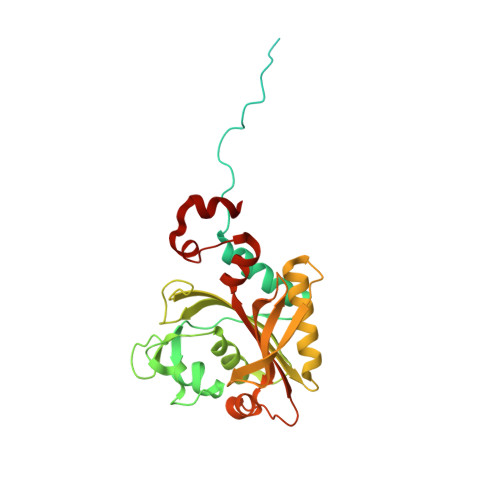
The role of a novel auxiliary pocket in bacterial phenylalanyl-tRNA synthetase druggability.
Abibi, A., Ferguson, A.D., Fleming, P.R., Gao, N., Hajec, L.I., Hu, J., Laganas, V.A., McKinney, D.C., McLeod, S.M., Prince, D.B., Shapiro, A.B., Buurman, E.T.(2014) J Biological Chem 289: 21651-21662
- PubMed: 24936059
- DOI: https://doi.org/10.1074/jbc.M114.574061
- Primary Citation of Related Structures:
4P71, 4P72, 4P73, 4P74, 4P75 - PubMed Abstract:
The antimicrobial activity of phenyl-thiazolylurea-sulfonamides against Staphylococcus aureus PheRS are dependent upon phenylalanine levels in the extracellular fluids. Inhibitor efficacy in animal models of infection is substantially diminished by dietary phenylalanine intake, thereby reducing the perceived clinical utility of this inhibitor class. The search for novel antibacterial compounds against Gram-negative pathogens led to a re-evaluation of this phenomenon, which is shown here to be unique to S. aureus. Inhibition of macromolecular syntheses and characterization of novel resistance mutations in Escherichia coli demonstrate that antimicrobial activity of phenyl-thiazolylurea-sulfonamides is mediated by PheRS inhibition, validating this enzyme as a viable drug discovery target for Gram-negative pathogens. A search for novel inhibitors of PheRS yielded three novel chemical starting points. NMR studies were used to confirm direct target engagement for phenylalanine-competitive hits. The crystallographic structure of Pseudomonas aeruginosa PheRS defined the binding modes of these hits and revealed an auxiliary hydrophobic pocket that is positioned adjacent to the phenylalanine binding site. Three viable inhibitor-resistant mutants were mapped to this pocket, suggesting that this region is a potential liability for drug discovery.
- From the Departments of Biosciences and.
Organizational Affiliation: