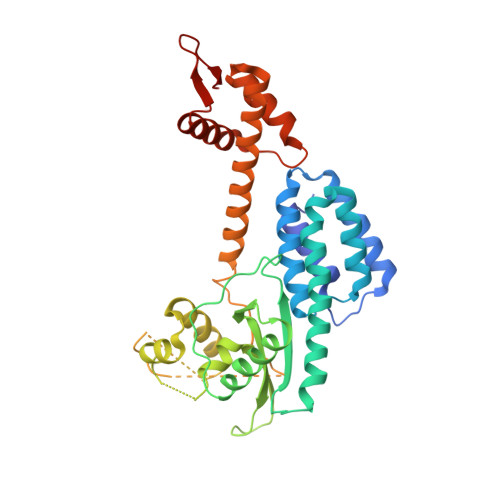
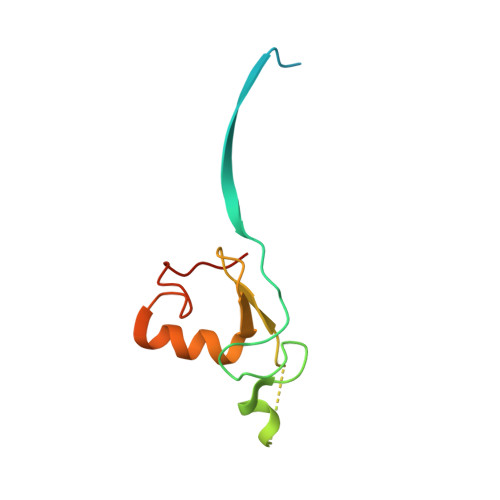
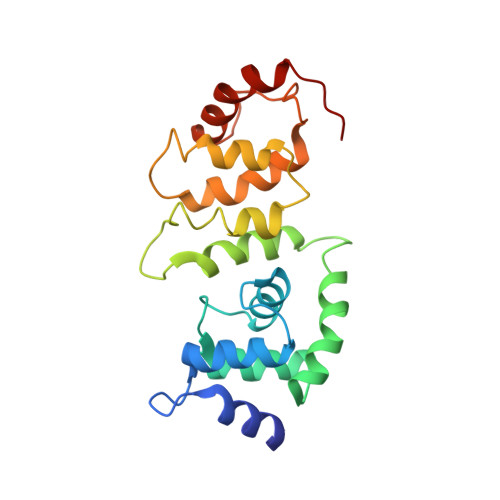
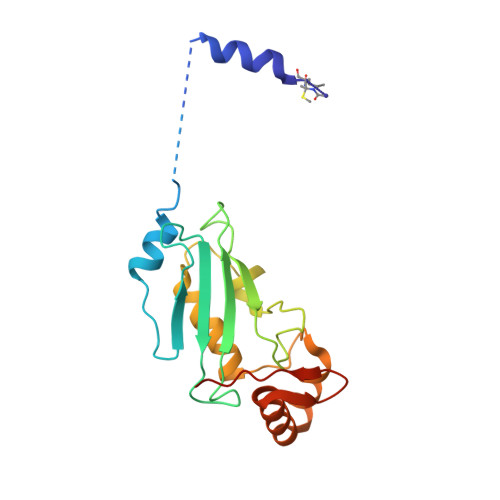
Structure of a RING E3 Trapped in Action Reveals Ligation Mechanism for the Ubiquitin-like Protein NEDD8.
Scott, D.C., Sviderskiy, V.O., Monda, J.K., Lydeard, J.R., Cho, S.E., Harper, J.W., Schulman, B.A.(2014) Cell 157: 1671-1684
- PubMed: 24949976
- DOI: https://doi.org/10.1016/j.cell.2014.04.037
- Primary Citation of Related Structures:
4P5O - PubMed Abstract:
Most E3 ligases use a RING domain to activate a thioester-linked E2∼ubiquitin-like protein (UBL) intermediate and promote UBL transfer to a remotely bound target protein. Nonetheless, RING E3 mechanisms matching a specific UBL and acceptor lysine remain elusive, including for RBX1, which mediates NEDD8 ligation to cullins and >10% of all ubiquitination. We report the structure of a trapped RING E3-E2∼UBL-target intermediate representing RBX1-UBC12∼NEDD8-CUL1-DCN1, which reveals the mechanism of NEDD8 ligation and how a particular UBL and acceptor lysine are matched by a multifunctional RING E3. Numerous mechanisms specify cullin neddylation while preventing noncognate ubiquitin ligation. Notably, E2-E3-target and RING-E2∼UBL modules are not optimized to function independently, but instead require integration by the UBL and target for maximal reactivity. The UBL and target regulate the catalytic machinery by positioning the RING-E2∼UBL catalytic center, licensing the acceptor lysine, and influencing E2 reactivity, thereby driving their specific coupling by a multifunctional RING E3.
- Department of Structural Biology, St. Jude Children's Research Hospital, Memphis, TN 38105, USA; Department of Tumor Cell Biology, St. Jude Children's Research Hospital, Memphis, TN 38105, USA; Howard Hughes Medical Institute, St. Jude Children's Research Hospital, Memphis, TN 38105, USA.
Organizational Affiliation: