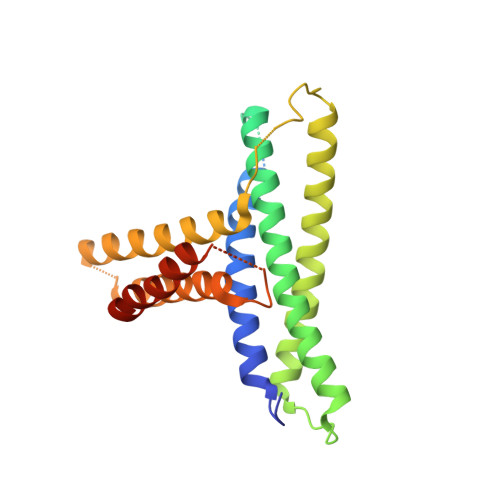
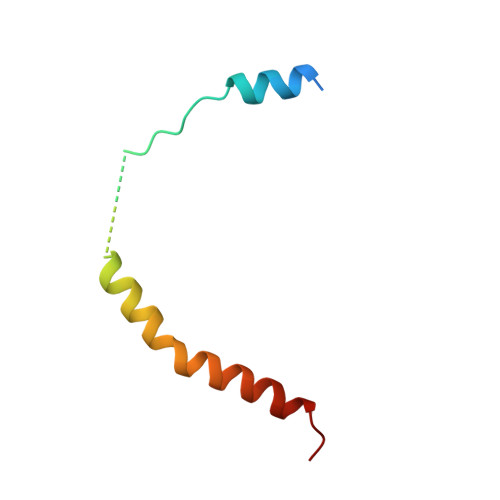
Structural basis of starvation-induced assembly of the autophagy initiation complex.
Fujioka, Y., Suzuki, S.W., Yamamoto, H., Kondo-Kakuta, C., Kimura, Y., Hirano, H., Akada, R., Inagaki, F., Ohsumi, Y., Noda, N.N.(2014) Nat Struct Mol Biol 21: 513-521
- PubMed: 24793651
- DOI: https://doi.org/10.1038/nsmb.2822
- Primary Citation of Related Structures:
4P1N, 4P1W - PubMed Abstract:
Assembly of the preautophagosomal structure (PAS) is essential for autophagy initiation in yeast. Starvation-induced dephosphorylation of Atg13 is required for the formation of the Atg1-Atg13-Atg17-Atg29-Atg31 complex (Atg1 complex), a prerequisite for PAS assembly. However, molecular details underlying these events have not been established. Here we studied the interactions of yeast Atg13 with Atg1 and Atg17 by X-ray crystallography. Atg13 binds tandem microtubule interacting and transport domains in Atg1, using an elongated helix-loop-helix region. Atg13 also binds Atg17, using a short region, thereby bridging Atg1 and Atg17 and leading to Atg1-complex formation. Dephosphorylation of specific serines in Atg13 enhanced its interaction with not only Atg1 but also Atg17. These observations update the autophagy-initiation model as follows: upon starvation, dephosphorylated Atg13 binds both Atg1 and Atg17, and this promotes PAS assembly and autophagy progression.
- 1] Institute of Microbial Chemistry (BIKAKEN), Tokyo, Japan. [2].
Organizational Affiliation: