Methanobactin production by methanotrophic bacteria and their structural diversity from Methylosinus strains: Insights into copper release
El Ghazouani, A., Basle, A., Dennison, C.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Find similar proteins by: Sequence | 3D Structure
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Methanobactin | 10 | Methylosinus sporium | Mutation(s): 0 | 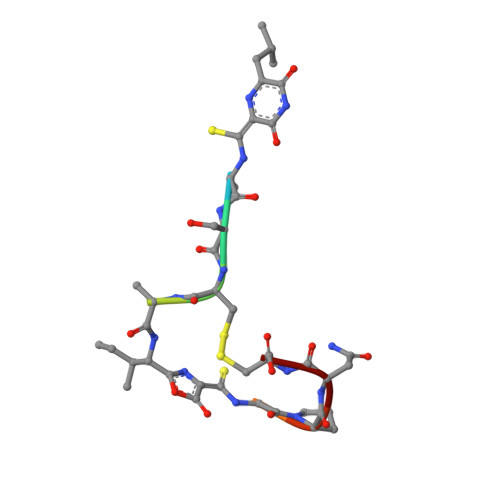 | |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| CU1 Query on CU1 | C [auth A], D [auth B] | COPPER (I) ION Cu VMQMZMRVKUZKQL-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 36.72 | α = 90 |
| b = 39.42 | β = 90 |
| c = 40.24 | γ = 90 |
| Software Name | Purpose |
|---|---|
| REFMAC | refinement |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Natural Environment Research Council (NERC) | United Kingdom | NE/F00608X |