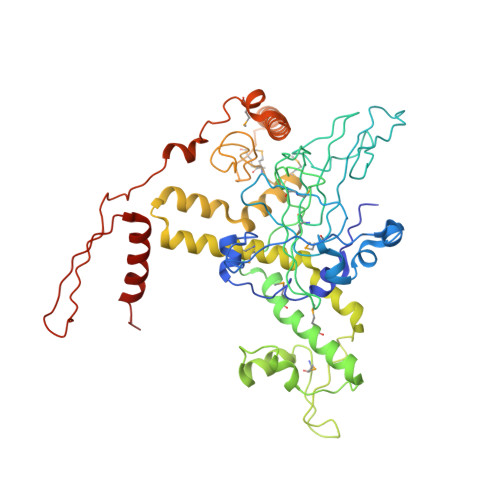
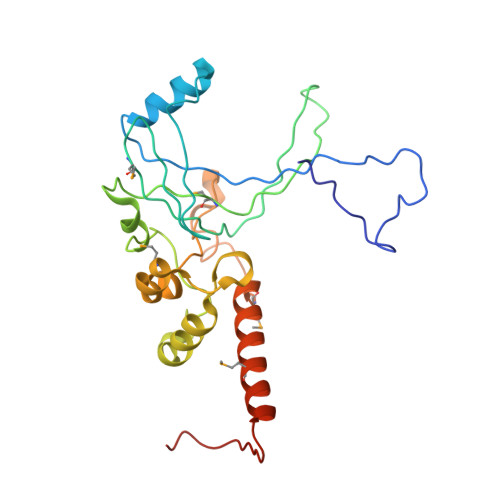
Structural and functional insight into TAF1-TAF7, a subcomplex of transcription factor II D.
Bhattacharya, S., Lou, X., Hwang, P., Rajashankar, K.R., Wang, X., Gustafsson, J.-A., Fletterick, R.J., Jacobson, R.H., Webb, P.(2014) Proc Natl Acad Sci U S A 111: 9103-9108
- PubMed: 24927529
- DOI: https://doi.org/10.1073/pnas.1408293111
- Primary Citation of Related Structures:
4OY2 - PubMed Abstract:
Transcription factor II D (TFIID) is a multiprotein complex that nucleates formation of the basal transcription machinery. TATA binding protein-associated factors 1 and 7 (TAF1 and TAF7), two subunits of TFIID, are integral to the regulation of eukaryotic transcription initiation and play key roles in preinitiation complex (PIC) assembly. Current models suggest that TAF7 acts as a dissociable inhibitor of TAF1 histone acetyltransferase activity and that this event ensures appropriate assembly of the RNA polymerase II-mediated PIC before transcriptional initiation. Here, we report the 3D structure of a complex of yeast TAF1 with TAF7 at 2.9 Å resolution. The structure displays novel architecture and is characterized by a large predominantly hydrophobic heterodimer interface and extensive cofolding of TAF subunits. There are no obvious similarities between TAF1 and known histone acetyltransferases. Instead, the surface of the TAF1-TAF7 complex contains two prominent conserved surface pockets, one of which binds selectively to an inhibitory trimethylated histone H3 mark on Lys27 in a manner that is also regulated by phosphorylation at the neighboring H3 serine. Our findings could point toward novel roles for the TAF1-TAF7 complex in regulation of PIC assembly via reading epigenetic histone marks.
- Genomic Medicine Program, Houston Methodist Research Institute, Houston, TX 77030;
Organizational Affiliation: