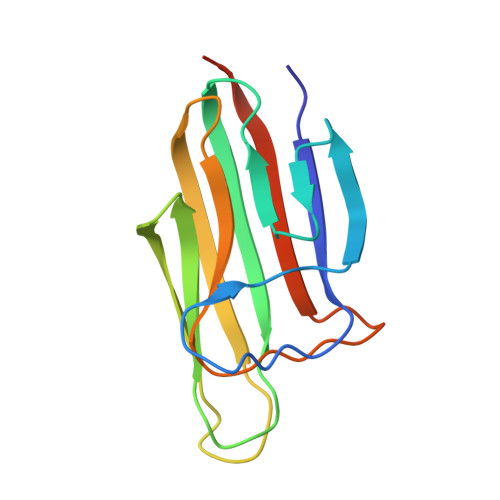
Structural insights into the C1q domain of Caprin-2 in canonical Wnt signaling
Miao, H., Jia, Y., Xie, S., Wang, X., Zhao, J., Chu, Y., Zhou, Z., Shi, Z., Song, X., Li, L.(2014) J Biological Chem 289: 34104-34113
- PubMed: 25331957
- DOI: https://doi.org/10.1074/jbc.M114.591636
- Primary Citation of Related Structures:
4OUL, 4OUM, 4OUS - PubMed Abstract:
Previously, we have identified Caprin-2 as a new regulator in canonical Wnt signaling through a mechanism of facilitating LRP5/6 phosphorylation; moreover, we found that its C-terminal C1q-related domain (Cap2_CRD) is required for this process. Here, we determined the crystal structures of Cap2_CRD from human and zebrafish, which both associate as a homotrimer with calcium located at the symmetric center. Surprisingly, the calcium binding-deficient mutant exists as a more stable trimer than its wild-type counterpart. Further studies showed that this Caprin-2 mutant disabled in binding calcium maintains the activity of promoting LRP5/6 phosphorylation, whereas the mutations disrupting Cap2_CRD homotrimer did impair such activity. Together, our findings suggested that the C-terminal CRD domain of Caprin-2 forms a flexible homotrimer mediated by calcium and that such trimeric assembly is required for Caprin-2 to regulate canonical Wnt signaling.
- From the State Key Laboratory of Molecular Biology, Institute of Biochemistry and Cell Biology, Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences, Shanghai 200031, China.
Organizational Affiliation: