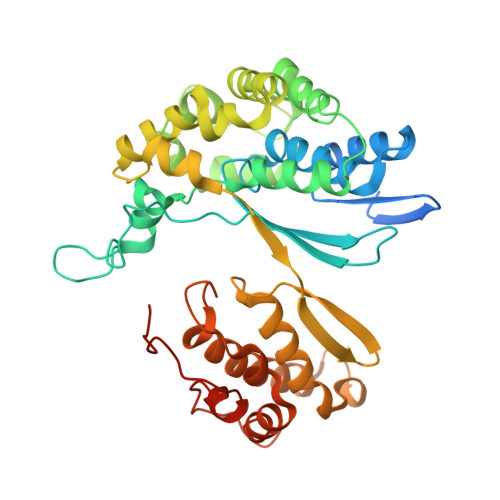
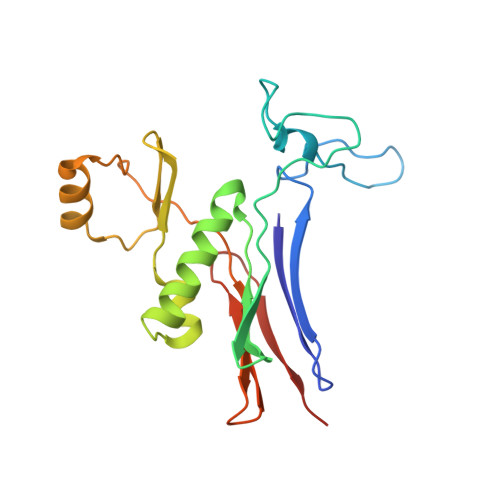
Low resolution X-ray structure of gamma-glutamyltranspeptidase from Bacillus licheniformis: Opened active site cleft and a cluster of acid residues potentially involved in the recognition of a metal ion.
Lin, L.L., Chen, Y.Y., Chi, M.C., Merlino, A.(2014) Biochim Biophys Acta 1844: 1523-1529
- PubMed: 24780583
- DOI: https://doi.org/10.1016/j.bbapap.2014.04.016
- Primary Citation of Related Structures:
4OTT, 4OTU - PubMed Abstract:
γ-Glutamyltranspeptidases (γ-GTs) cleave the γ-glutamyl amide bond of glutathione and transfer the released γ-glutamyl group to water (hydrolysis) or acceptor amino acids (transpeptidation). These ubiquitous enzymes play a key role in the biosynthesis and degradation of glutathione, and in xenobiotic detoxification. Here we report the 3Å resolution crystal structure of Bacillus licheniformis γ-GT (BlGT) and that of its complex with l-Glu. X-ray structures confirm that BlGT belongs to the N-terminal nucleophilic hydrolase superfamily and reveal that the protein possesses an opened active site cleft similar to that reported for the homologous enzyme from Bacillus subtilis, but different from those observed for human γ-GT and for γ-GTs from other microorganisms. Data suggest that the binding of l-Glu induces a reordering of the C-terminal tail of BlGT large subunit and allow the identification of a cluster of acid residues that are potentially involved in the recognition of a metal ion. The role of these residues on the conformational stability of BlGT has been studied by characterizing the autoprocessing, enzymatic activity, chemical and thermal denaturation of four new Ala single mutants. The results show that replacement of Asp568 with an Ala affects both the autoprocessing and structural stability of the protein.
- Department of Applied Chemistry, National Chiayi University, 300 Syuefu Road, Chiayi City 600, Taiwan.
Organizational Affiliation: