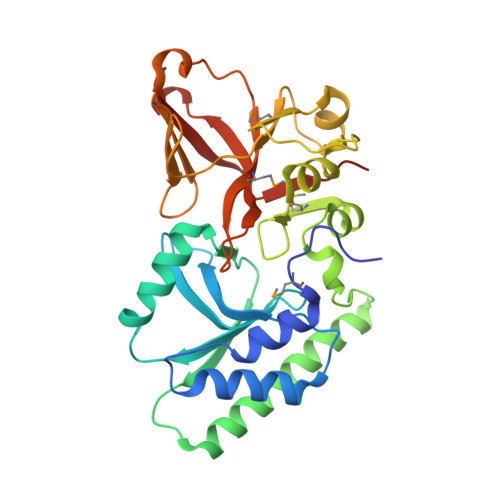
Crystal structure of the 5hmC specific endonuclease PvuRts1I.
Kazrani, A.A., Kowalska, M., Czapinska, H., Bochtler, M.(2014) Nucleic Acids Res 42: 5929-5936
- PubMed: 24634440
- DOI: https://doi.org/10.1093/nar/gku186
- Primary Citation of Related Structures:
4OQ2 - PubMed Abstract:
PvuRts1I is a prototype for a larger family of restriction endonucleases that cleave DNA containing 5-hydroxymethylcytosine (5hmC) or 5-glucosylhydroxymethylcytosine (5ghmC), but not 5-methylcytosine (5mC) or cytosine. Here, we report a crystal structure of the enzyme at 2.35 Å resolution. Although the protein has been crystallized in the absence of DNA, the structure is very informative. It shows that PvuRts1I consists of an N-terminal, atypical PD-(D/E)XK catalytic domain and a C-terminal SRA domain that might accommodate a flipped 5hmC or 5ghmC base. Changes to predicted catalytic residues of the PD-(D/E)XK domain or to the putative pocket for a flipped base abolish catalytic activity. Surprisingly, fluorescence changes indicative of base flipping are not observed when PvuRts1I is added to DNA substrates containing pyrrolocytosine in place of 5hmC (5ghmC). Despite this caveat, the structure suggests a model for PvuRts1I activity and presents opportunities for protein engineering to alter the enzyme properties for biotechnological applications.
- International Institute of Molecular and Cell Biology, Trojdena 4, 02109 Warsaw, Poland mbochtler@iimcb.gov.pl.
Organizational Affiliation: