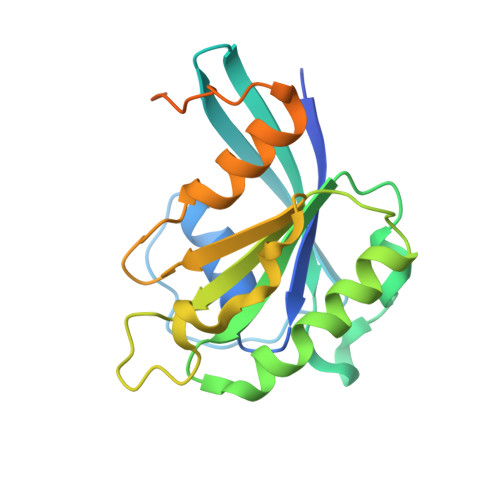
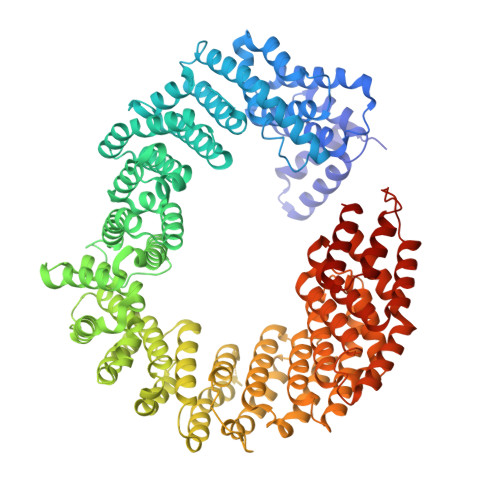
Structure of transportin SR2, a karyopherin involved in human disease, in complex with Ran.
Tsirkone, V.G., Beutels, K.G., Demeulemeester, J., Debyser, Z., Christ, F., Strelkov, S.V.(2014) Acta Crystallogr Sect F Struct Biol Cryst Commun 70: 723-729
- PubMed: 24915079
- DOI: https://doi.org/10.1107/S2053230X14009492
- Primary Citation of Related Structures:
4OL0 - PubMed Abstract:
Transportin SR2 (TRN-SR2) is a β-type karyopherin responsible for the nuclear import of specific cargoes, including serine/arginine-rich splicing factors. The protein has been implicated in a variety of human diseases, including HIV infection, primary biliary cirrhosis and limb-girdle muscular dystrophy 1F. Towards understanding its molecular mechanism, a 2.9 Å resolution crystal structure of human TRN-SR2 complexed with the small GTPase Ran has been determined. TRN-SR2 is composed of 20 α-helical HEAT repeats forming a solenoid-like fold. The first nine repeats form a `cradle' for the binding of RanGTP, revealing similarities but also differences with respect to the related importin 13 complex.
- Laboratory for Biocrystallography, Department of Pharmaceutical and Pharmacological Sciences, KU Leuven, Herestraat 49 bus 822, 3000 Leuven, Belgium.
Organizational Affiliation: