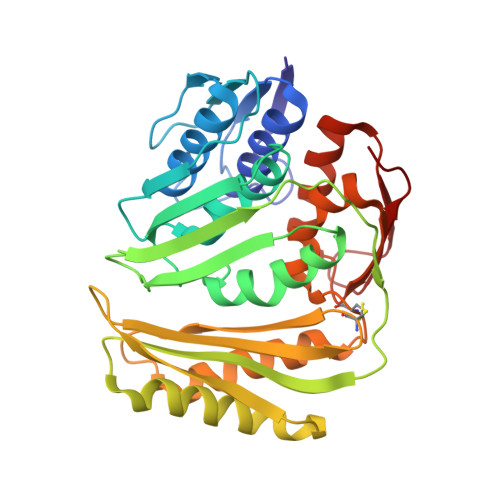
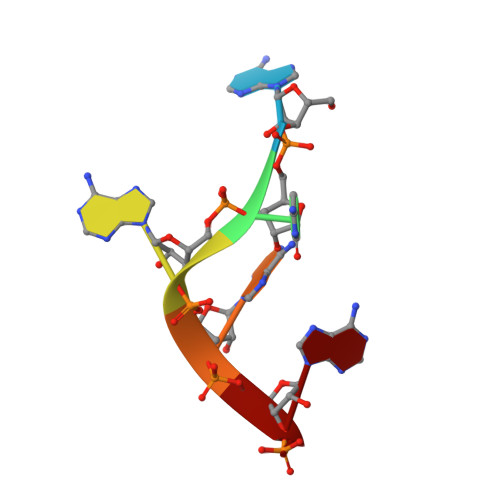
Structure of RNA 3'-phosphate cyclase bound to substrate RNA.
Desai, K.K., Bingman, C.A., Cheng, C.L., Phillips Jr., G.N., Raines, R.T.(2014) RNA 20: 1560-1566
- PubMed: 25161314
- DOI: https://doi.org/10.1261/rna.045823.114
- Primary Citation of Related Structures:
4O89, 4O8J - PubMed Abstract:
RNA 3'-phosphate cyclase (RtcA) catalyzes the ATP-dependent cyclization of a 3'-phosphate to form a 2',3'-cyclic phosphate at RNA termini. Cyclization proceeds through RtcA-AMP and RNA(3')pp(5')A covalent intermediates, which are analogous to intermediates formed during catalysis by the tRNA ligase RtcB. Here we present a crystal structure of Pyrococcus horikoshii RtcA in complex with a 3'-phosphate terminated RNA and adenosine in the AMP-binding pocket. Our data reveal that RtcA recognizes substrate RNA by ensuring that the terminal 3'-phosphate makes a large contribution to RNA binding. Furthermore, the RNA 3'-phosphate is poised for in-line attack on the P-N bond that links the phosphorous atom of AMP to N(ε) of His307. Thus, we provide the first insights into RNA 3'-phosphate termini recognition and the mechanism of 3'-phosphate activation by an Rtc enzyme.
- Department of Biochemistry, University of Wisconsin-Madison, Madison, Wisconsin 53706, USA.
Organizational Affiliation: