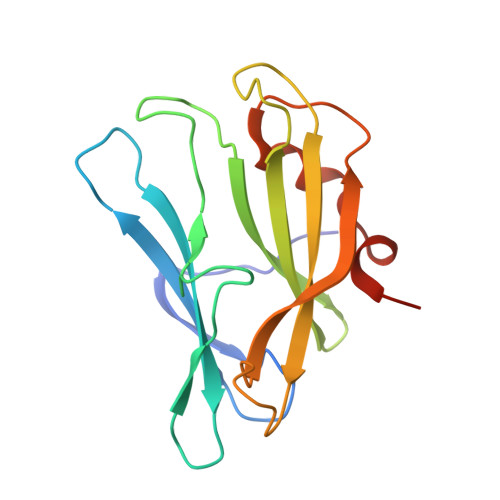
Crystal structure of the carbapenem intrinsic resistance protein CarG
Tichy, E.M., Luisi, B.F., Salmond, G.P.C.(2014) J Mol Biology 426: 1958-1970
- PubMed: 24583229
- DOI: https://doi.org/10.1016/j.jmb.2014.02.016
- Primary Citation of Related Structures:
4O7J - PubMed Abstract:
In the Gram-negative enterobacterium Erwinia (Pectobacterium) and Serratia sp. ATCC 39006, intrinsic resistance to the carbapenem antibiotic 1-carbapen-2-em-3-carboxylic acid is mediated by the CarF and CarG proteins, by an unknown mechanism. Here, we report a high-resolution crystal structure for the Serratia sp. ATCC 39006 carbapenem resistance protein CarG. This structure of CarG is the first in the carbapenem intrinsic resistance (CIR) family of resistance proteins from carbapenem-producing bacteria. The crystal structure shows the protein to form a homodimer, in agreement with results from analytical gel filtration. The structure of CarG does not show homology with any known antibiotic resistance proteins nor does it belong to any well-characterised protein structural family. However, it is a close structural homologue of the bacterial inhibitor of invertebrate lysozyme, PliI-Ah, with some interesting structural variations, including the absence of the catalytic site responsible for lysozyme inhibition. Both proteins show a unique β-sandwich fold with short terminal α-helices. The core of the protein is formed by stacked anti-parallel sheets that are individually very similar in the two proteins but differ in their packing interface, causing the splaying of the two sheets in CarG. Furthermore, a conserved cation binding site identified in CarG is absent from the homologue.
- Department of Biochemistry, University of Cambridge, Building O, Downing Site, Cambridge CB21QW, UK.
Organizational Affiliation: