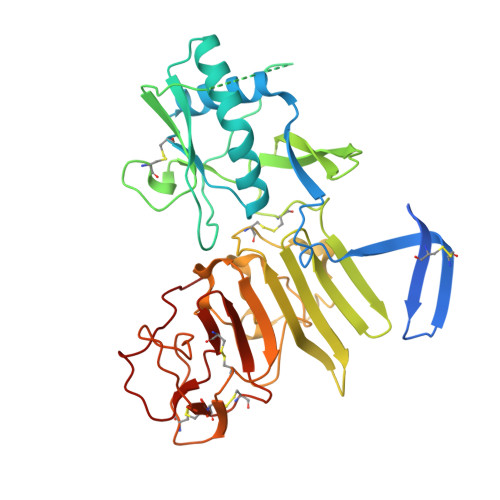
Flavivirus NS1 structures reveal surfaces for associations with membranes and the immune system.
Akey, D.L., Brown, W.C., Dutta, S., Konwerski, J., Jose, J., Jurkiw, T.J., DelProposto, J., Ogata, C.M., Skiniotis, G., Kuhn, R.J., Smith, J.L.(2014) Science 343: 881-885
- PubMed: 24505133
- DOI: https://doi.org/10.1126/science.1247749
- Primary Citation of Related Structures:
4O6B, 4O6C, 4O6D - PubMed Abstract:
Flaviviruses, the human pathogens responsible for dengue fever, West Nile fever, tick-borne encephalitis, and yellow fever, are endemic in tropical and temperate parts of the world. The flavivirus nonstructural protein 1 (NS1) functions in genome replication as an intracellular dimer and in immune system evasion as a secreted hexamer. We report crystal structures for full-length, glycosylated NS1 from West Nile and dengue viruses. The NS1 hexamer in crystal structures is similar to a solution hexamer visualized by single-particle electron microscopy. Recombinant NS1 binds to lipid bilayers and remodels large liposomes into lipoprotein nanoparticles. The NS1 structures reveal distinct domains for membrane association of the dimer and interactions with the immune system and are a basis for elucidating the molecular mechanism of NS1 function.
- Life Sciences Institute, University of Michigan, Ann Arbor, MI 48109, USA.
Organizational Affiliation: