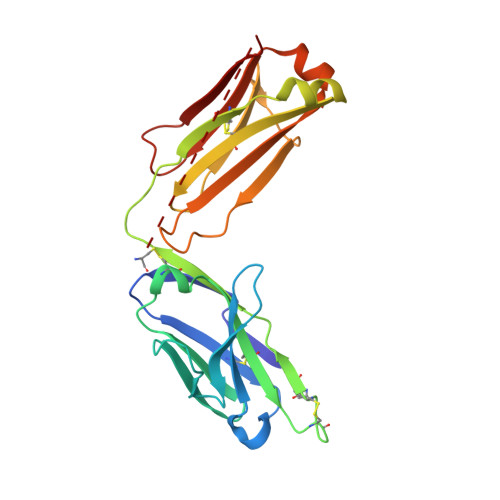
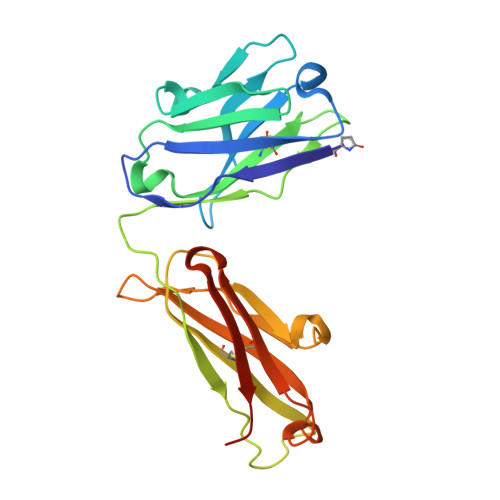
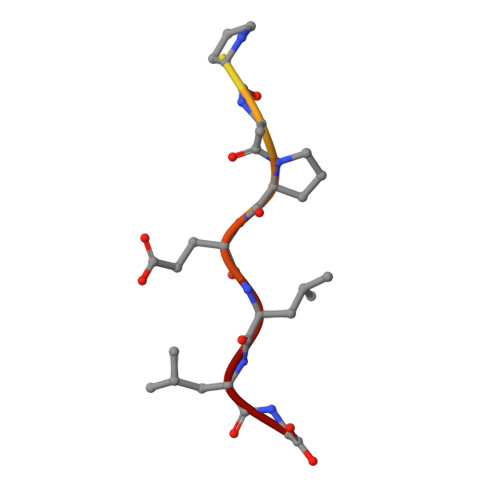
Structure and specificity of an antibody targeting a proteolytically cleaved IgG hinge.
Malia, T.J., Teplyakov, A., Brezski, R.J., Luo, J., Kinder, M., Sweet, R.W., Almagro, J.C., Jordan, R.E., Gilliland, G.L.(2014) Proteins 82: 1656-1667
- PubMed: 24638881
- DOI: https://doi.org/10.1002/prot.24545
- Primary Citation of Related Structures:
4MA3, 4O4Y, 4O51 - PubMed Abstract:
The functional role of human antihinge (HAH) autoantibodies in normal health and disease remains elusive, but recent evidence supports their role in the host response to IgG cleavage by proteases that are prevalent in certain disorders. Characterization and potential exploitation of these HAH antibodies has been hindered by the absence of monoclonal reagents. 2095-2 is a rabbit monoclonal antibody targeting the IdeS-cleaved hinge of human IgG1. We have determined the crystal structure of the Fab of 2095-2 and its complex with a hinge analog peptide. The antibody is selective for the C-terminally cleaved hinge ending in G236 and this interaction involves an uncommon disulfide in VL CDR3. We probed the importance of the disulfide in VL CDR3 through engineering variants. We identified one variant, QAA, which does not require the disulfide for biological activity or peptide binding. The structure of this variant offers a starting point for further engineering of 2095-2 with the same specificity, but lacking the potential manufacturing liability of an additional disulfide. Proteins 2014; 82:1656-1667. © 2014 Wiley Periodicals, Inc.
- Biologics Research, Janssen Research and Development, LLC, Spring House, Pennsylvania.
Organizational Affiliation: