Structure of L-isoaspartyl (D-aspartyl) Methyltransferase
Griffith, S.C.(2002) Thesis University of California, Los Angeles: 93-107
Experimental Data Snapshot
Starting Model: experimental
View more details
(2002) Thesis University of California, Los Angeles: 93-107
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Protein-L-isoaspartate O-methyltransferase | 208 | Pyrobaculum aerophilum str. IM2 | Mutation(s): 0 Gene Names: PAE0701, pcm, PIMT EC: 2.1.1.77 | 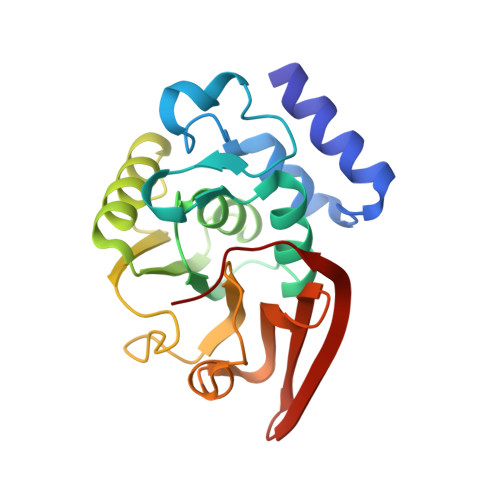 | |
UniProt | |||||
Find proteins for Q8ZYN0 (Pyrobaculum aerophilum (strain ATCC 51768 / DSM 7523 / JCM 9630 / CIP 104966 / NBRC 100827 / IM2)) Explore Q8ZYN0 Go to UniProtKB: Q8ZYN0 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q8ZYN0 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| SAH Query on SAH | B [auth A] | S-ADENOSYL-L-HOMOCYSTEINE C14 H20 N6 O5 S ZJUKTBDSGOFHSH-WFMPWKQPSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 73.72 | α = 90 |
| b = 35.67 | β = 100.31 |
| c = 66.29 | γ = 90 |
| Software Name | Purpose |
|---|---|
| XSCALE | data scaling |
| EPMR | phasing |
| BUSTER-TNT | refinement |
| PDB_EXTRACT | data extraction |
| BUSTER | refinement |