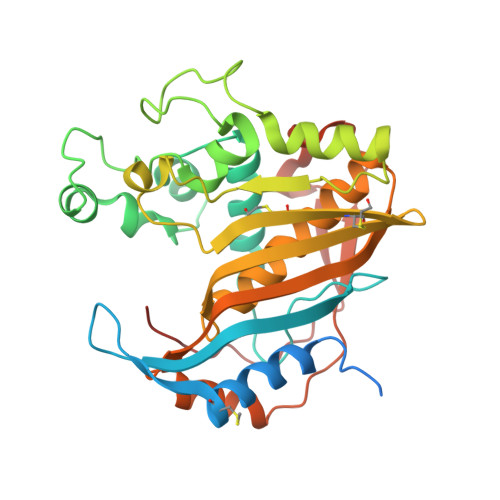
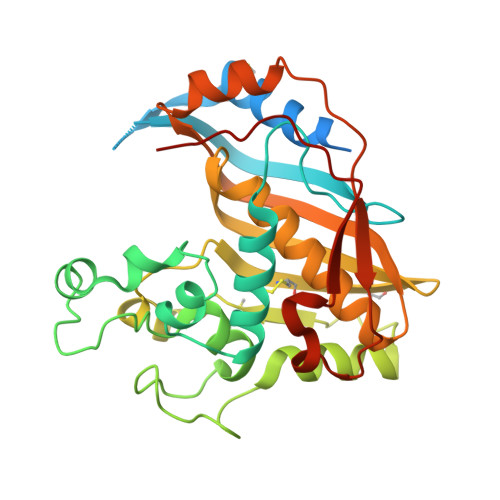
Destabilizers of the thymidylate synthase homodimer accelerate its proteasomal degradation and inhibit cancer growth.
Costantino, L., Ferrari, S., Santucci, M., Salo-Ahen, O.M.H., Carosati, E., Franchini, S., Lauriola, A., Pozzi, C., Trande, M., Gozzi, G., Saxena, P., Cannazza, G., Losi, L., Cardinale, D., Venturelli, A., Quotadamo, A., Linciano, P., Tagliazucchi, L., Moschella, M.G., Guerrini, R., Pacifico, S., Luciani, R., Genovese, F., Henrich, S., Alboni, S., Santarem, N., da Silva Cordeiro, A., Giovannetti, E., Peters, G.J., Pinton, P., Rimessi, A., Cruciani, G., Stroud, R.M., Wade, R.C., Mangani, S., Marverti, G., D'Arca, D., Ponterini, G., Costi, M.P.(2022) Elife 11
- PubMed: 36475542
- DOI: https://doi.org/10.7554/eLife.73862
- Primary Citation of Related Structures:
4O1U, 4O1X - PubMed Abstract:
Drugs that target human thymidylate synthase (hTS), a dimeric enzyme, are widely used in anticancer therapy. However, treatment with classical substrate-site-directed TS inhibitors induces over-expression of this protein and development of drug resistance. We thus pursued an alternative strategy that led us to the discovery of TS-dimer destabilizers. These compounds bind at the monomer-monomer interface and shift the dimerization equilibrium of both the recombinant and the intracellular protein toward the inactive monomers. A structural, spectroscopic, and kinetic investigation has provided evidence and quantitative information on the effects of the interaction of these small molecules with hTS. Focusing on the best among them, E7 , we have shown that it inhibits hTS in cancer cells and accelerates its proteasomal degradation, thus causing a decrease in the enzyme intracellular level. E7 also showed a superior anticancer profile to fluorouracil in a mouse model of human pancreatic and ovarian cancer. Thus, over sixty years after the discovery of the first TS prodrug inhibitor, fluorouracil, E7 breaks the link between TS inhibition and enhanced expression in response, providing a strategy to fight drug-resistant cancers.
- Department of Life Sciences, University of Modena and Reggio Emilia, Modena, Italy.
Organizational Affiliation: