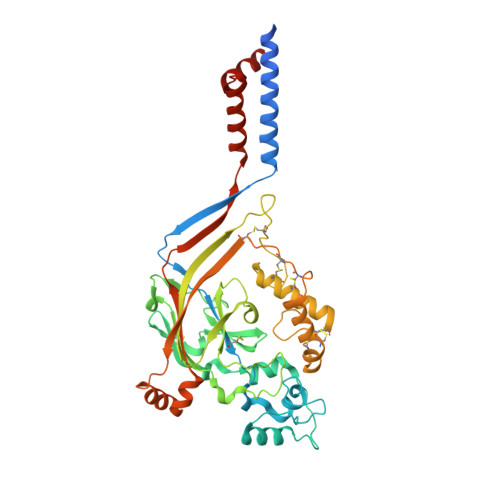
X-ray structure of Acid-sensing ion channel 1-snake toxin complex reveals open state of a na(+)-selective channel.
Baconguis, I., Bohlen, C.J., Goehring, A., Julius, D., Gouaux, E.(2014) Cell 156: 717-729
- PubMed: 24507937
- DOI: https://doi.org/10.1016/j.cell.2014.01.011
- Primary Citation of Related Structures:
4NTW, 4NTX, 4NTY - PubMed Abstract:
Acid-sensing ion channels (ASICs) detect extracellular protons produced during inflammation or ischemic injury and belong to the superfamily of degenerin/epithelial sodium channels. Here, we determine the cocrystal structure of chicken ASIC1a with MitTx, a pain-inducing toxin from the Texas coral snake, to define the structure of the open state of ASIC1a. In the MitTx-bound open state and in the previously determined low-pH desensitized state, TM2 is a discontinuous α helix in which the Gly-Ala-Ser selectivity filter adopts an extended, belt-like conformation, swapping the cytoplasmic one-third of TM2 with an adjacent subunit. Gly 443 residues of the selectivity filter provide a ring of three carbonyl oxygen atoms with a radius of ∼3.6 Å, presenting an energetic barrier for hydrated ions. The ASIC1a-MitTx complex illuminates the mechanism of MitTx action, defines the structure of the selectivity filter of voltage-independent, sodium-selective ion channels, and captures the open state of an ASIC.
- Vollum Institute, Oregon Health and Science University, 3181 SW Sam Jackson Park Road, Portland, OR 97239, USA.
Organizational Affiliation: