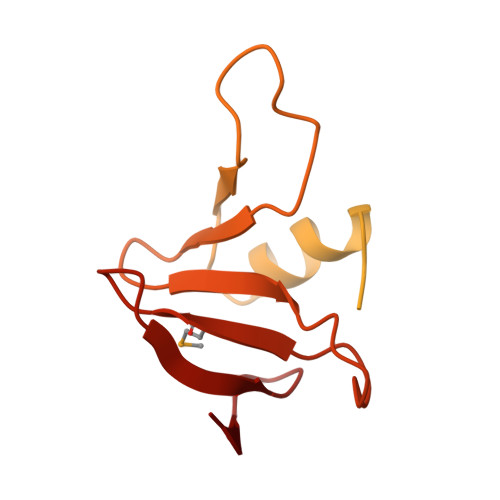
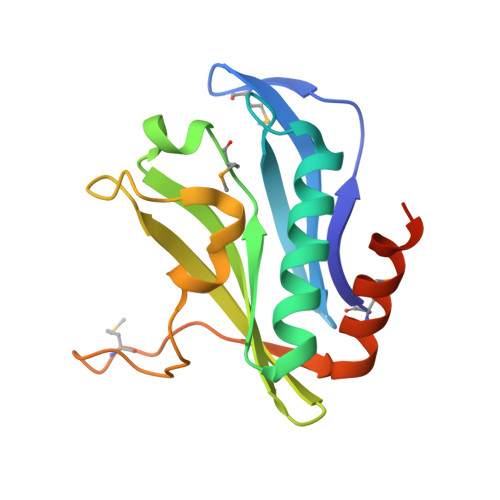
CdiA from Enterobacter cloacae Delivers a Toxic Ribosomal RNase into Target Bacteria.
Beck, C.M., Morse, R.P., Cunningham, D.A., Iniguez, A., Low, D.A., Goulding, C.W., Hayes, C.S.(2014) Structure 22: 1-12
- PubMed: 24657090
- DOI: https://doi.org/10.1016/j.str.2014.02.012
- Primary Citation of Related Structures:
4NTQ - PubMed Abstract:
Contact-dependent growth inhibition (CDI) is one mechanism of inter-bacterial competition. CDI(+) cells export large CdiA effector proteins, which carry a variety of C-terminal toxin domains (CdiA-CT). CdiA-CT toxins are specifically neutralized by cognate CdiI immunity proteins to protect toxin-producing cells from autoinhibition. Here, we use structure determination to elucidate the activity of a CDI toxin from Enterobacter cloacae (ECL). The structure of CdiA-CT(ECL) resembles the C-terminal nuclease domain of colicin E3, which cleaves 16S ribosomal RNA to disrupt protein synthesis. In accord with this structural homology, we show that CdiA-CT(ECL) uses the same nuclease activity to inhibit bacterial growth. Surprisingly, although colicin E3 and CdiA(ECL) carry equivalent toxin domains, the corresponding immunity proteins are unrelated in sequence, structure, and toxin-binding site. Together, these findings reveal unexpected diversity among 16S rRNases and suggest that these nucleases are robust and versatile payloads for a variety of toxin-delivery platforms.
- Department of Molecular, Cellular, and Developmental Biology, University of California, Santa Barbara, Santa Barbara, CA 93106-9625, USA.
Organizational Affiliation: