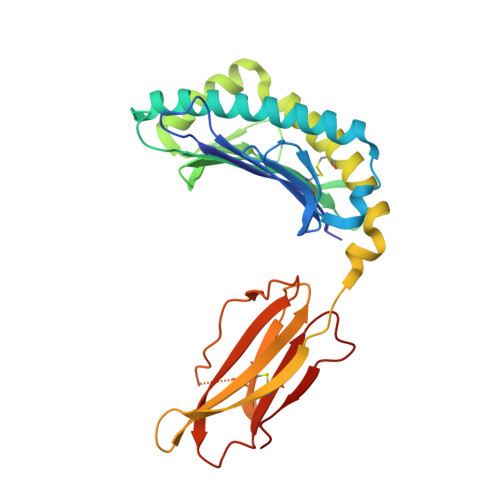
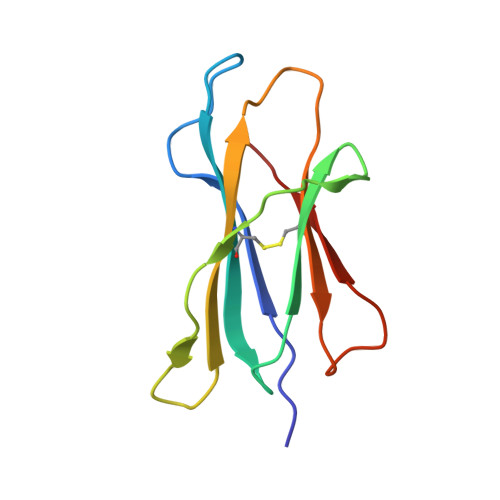
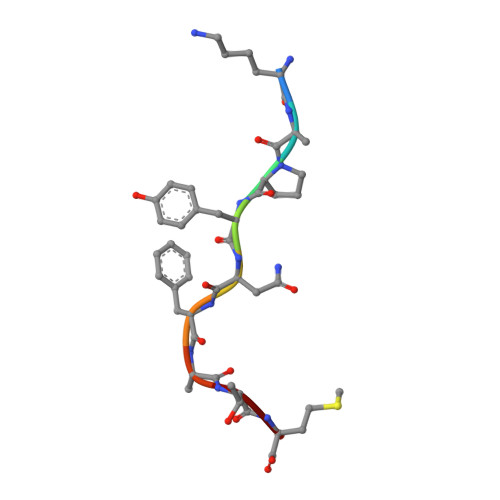
CRYSTAL STRUCTURE OF THE MURINE CLASS I MAJOR HISTOCOMPATIBILITY COMPLEX H-2DB IN COMPLEX WITH LCMV-DERIVED GP33 ALTERED PEPTIDE ligand V3P
Allerbring, E.B., Duru, A.D., Uchtenhagen, H., Mazumdar, P.A., Badia-Martinez, D., Madhurantakam, C., Sandalova, T., Nygren, P., Achour, A.To be published.