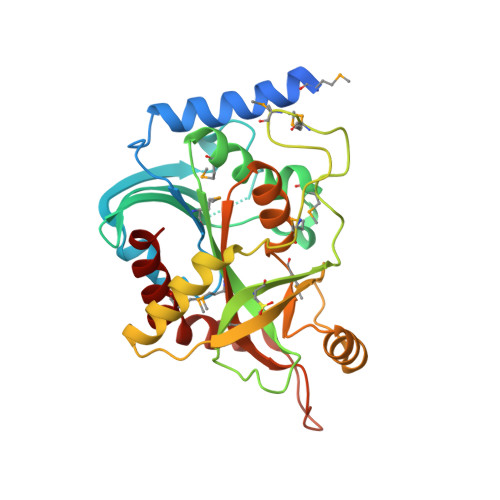
Crystal structure of purine nucleoside phosphorylase from Porphyromonas gingivalis ATCC 33277, NYSGRC Target 30972.
Malashkevich, V.N., Bhosle, R., Toro, R., Hillerich, B., Gizzi, A., Garforth, S., Kar, A., Chan, M.K., Lafluer, J., Patel, H., Matikainen, B., Chamala, S., Lim, S., Celikgil, A., Villegas, G., Evans, B., Love, J., Fiser, A., Seidel, R., Bonanno, J.B., Almo, S.C.To be published.