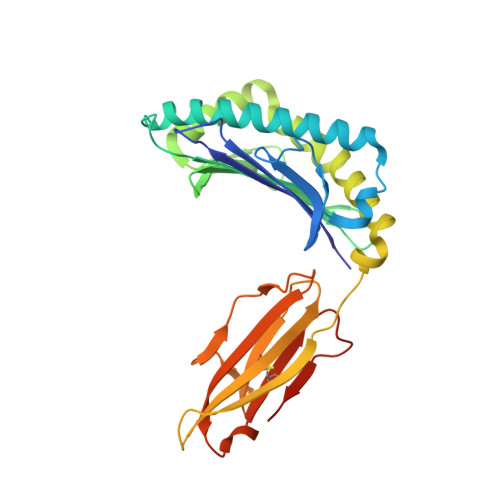
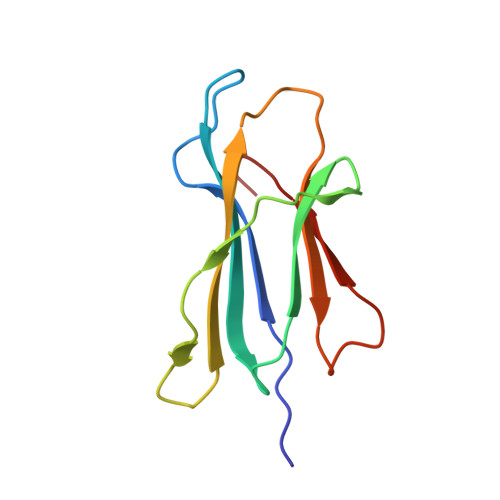
Preexisting CD8+ T-cell immunity to the H7N9 influenza A virus varies across ethnicities.
Quinones-Parra, S., Grant, E., Loh, L., Nguyen, T.H., Campbell, K.A., Tong, S.Y., Miller, A., Doherty, P.C., Vijaykrishna, D., Rossjohn, J., Gras, S., Kedzierska, K.(2014) Proc Natl Acad Sci U S A 111: 1049-1054
- PubMed: 24395804
- DOI: https://doi.org/10.1073/pnas.1322229111
- Primary Citation of Related Structures:
4NQV, 4NQX - PubMed Abstract:
The absence of preexisting neutralizing antibodies specific for the novel A (H7N9) influenza virus indicates a lack of prior human exposure. As influenza A virus-specific CD8(+) T lymphocytes (CTLs) can be broadly cross-reactive, we tested whether immunogenic peptides derived from H7N9 might be recognized by memory CTLs established following infection with other influenza strains. Probing across multiple ethnicities, we identified 32 conserved epitopes derived from the nucleoprotein (NP) and matrix-1 (M1) proteins. These NP and M1 peptides are presented by HLAs prevalent in 16-57% of individuals. Remarkably, some HLA alleles (A*0201, A*0301, B*5701, B*1801, and B*0801) elicit robust CTL responses against any human influenza A virus, including H7N9, whereas ethnicities where HLA-A*0101, A*6801, B*1501, and A*2402 are prominent, show limited CTL response profiles. By this criterion, some groups, especially the Alaskan and Australian Indigenous peoples, would be particularly vulnerable to H7N9 infection. This dissection of CTL-mediated immunity to H7N9 thus suggests strategies for both vaccine delivery and development.
- Department of Microbiology and Immunology, The University of Melbourne, at the Peter Doherty Institute for Infection and Immunity, Victoria, 3010, Australia.
Organizational Affiliation: