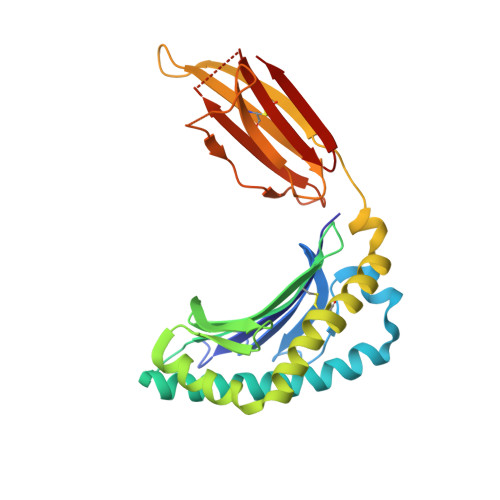
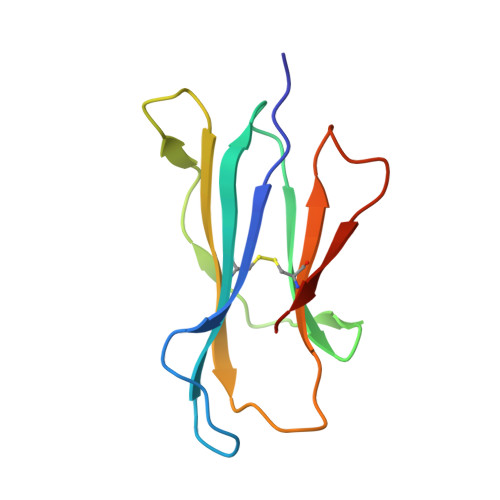
T-cell activation by transitory neo-antigens derived from distinct microbial pathways.
Corbett, A.J., Eckle, S.B., Birkinshaw, R.W., Liu, L., Patel, O., Mahony, J., Chen, Z., Reantragoon, R., Meehan, B., Cao, H., Williamson, N.A., Strugnell, R.A., Van Sinderen, D., Mak, J.Y., Fairlie, D.P., Kjer-Nielsen, L., Rossjohn, J., McCluskey, J.(2014) Nature 509: 361-365
- PubMed: 24695216
- DOI: https://doi.org/10.1038/nature13160
- Primary Citation of Related Structures:
4NQC, 4NQD, 4NQE - PubMed Abstract:
T cells discriminate between foreign and host molecules by recognizing distinct microbial molecules, predominantly peptides and lipids. Riboflavin precursors found in many bacteria and yeast also selectively activate mucosal-associated invariant T (MAIT) cells, an abundant population of innate-like T cells in humans. However, the genesis of these small organic molecules and their mode of presentation to MAIT cells by the major histocompatibility complex (MHC)-related protein MR1 (ref. 8) are not well understood. Here we show that MAIT-cell activation requires key genes encoding enzymes that form 5-amino-6-d-ribitylaminouracil (5-A-RU), an early intermediate in bacterial riboflavin synthesis. Although 5-A-RU does not bind MR1 or activate MAIT cells directly, it does form potent MAIT-activating antigens via non-enzymatic reactions with small molecules, such as glyoxal and methylglyoxal, which are derived from other metabolic pathways. The MAIT antigens formed by the reactions between 5-A-RU and glyoxal/methylglyoxal were simple adducts, 5-(2-oxoethylideneamino)-6-D-ribitylaminouracil (5-OE-RU) and 5-(2-oxopropylideneamino)-6-D-ribitylaminouracil (5-OP-RU), respectively, which bound to MR1 as shown by crystal structures of MAIT TCR ternary complexes. Although 5-OP-RU and 5-OE-RU are unstable intermediates, they became trapped by MR1 as reversible covalent Schiff base complexes. Mass spectra supported the capture by MR1 of 5-OP-RU and 5-OE-RU from bacterial cultures that activate MAIT cells, but not from non-activating bacteria, indicating that these MAIT antigens are present in a range of microbes. Thus, MR1 is able to capture, stabilize and present chemically unstable pyrimidine intermediates, which otherwise convert to lumazines, as potent antigens to MAIT cells. These pyrimidine adducts are microbial signatures for MAIT-cell immunosurveillance.
- 1] Department of Microbiology and Immunology, Peter Doherty Institute for Infection and Immunity, University of Melbourne, Parkville, Victoria 3010, Australia [2].
Organizational Affiliation: