Mechanism of Action and Epitopes of Clostridium difficile Toxin B-neutralizing Antibody Bezlotoxumab Revealed by X-ray Crystallography.
Orth, P., Xiao, L., Hernandez, L.D., Reichert, P., Sheth, P.R., Beaumont, M., Yang, X., Murgolo, N., Ermakov, G., DiNunzio, E., Racine, F., Karczewski, J., Secore, S., Ingram, R.N., Mayhood, T., Strickland, C., Therien, A.G.(2014) J Biological Chem 289: 18008-18021
- PubMed: 24821719
- DOI: https://doi.org/10.1074/jbc.M114.560748
- Primary Citation of Related Structures:
4NP4 - PubMed Abstract:
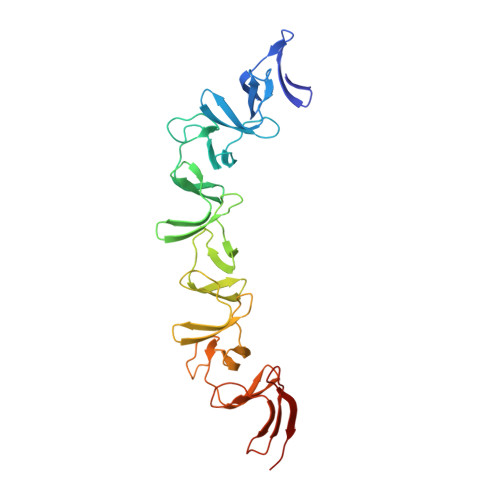
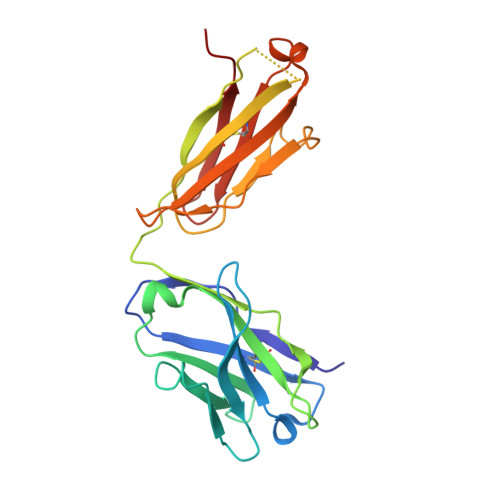
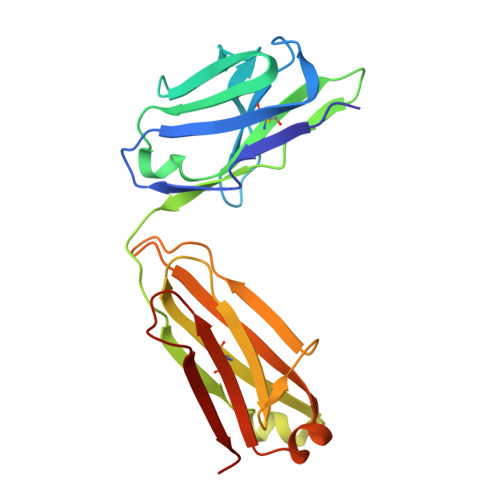
The symptoms of Clostridium difficile infections are caused by two exotoxins, TcdA and TcdB, which target host colonocytes by binding to unknown cell surface receptors, at least in part via their combined repetitive oligopeptide (CROP) domains. A combination of the anti-TcdA antibody actoxumab and the anti-TcdB antibody bezlotoxumab is currently under development for the prevention of recurrent C. difficile infections. We demonstrate here through various biophysical approaches that bezlotoxumab binds to specific regions within the N-terminal half of the TcdB CROP domain. Based on this information, we solved the x-ray structure of the N-terminal half of the TcdB CROP domain bound to Fab fragments of bezlotoxumab. The structure reveals that the TcdB CROP domain adopts a β-solenoid fold consisting of long and short repeats and that bezlotoxumab binds to two homologous sites within the CROP domain, partially occluding two of the four putative carbohydrate binding pockets located in TcdB. We also show that bezlotoxumab neutralizes TcdB by blocking binding of TcdB to mammalian cells. Overall, our data are consistent with a model wherein a single molecule of bezlotoxumab neutralizes TcdB by binding via its two Fab regions to two epitopes within the N-terminal half of the TcdB CROP domain, partially blocking the carbohydrate binding pockets of the toxin and preventing toxin binding to host cells.
- From Merck & Co., Inc., Kenilworth, New Jersey 07023.
Organizational Affiliation: