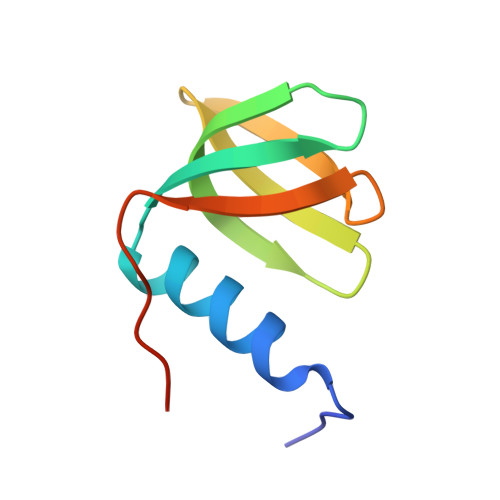
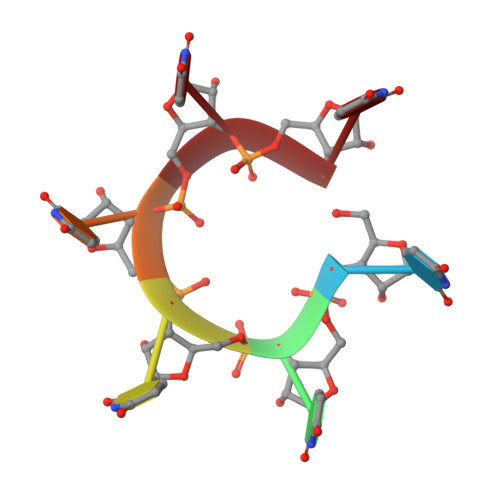
Recognition of U-rich RNA by Hfq from the Gram-positive pathogen Listeria monocytogenes.
Kovach, A.R., Hoff, K.E., Canty, J.T., Orans, J., Brennan, R.G.(2014) RNA 20: 1548-1559
- PubMed: 25150227
- DOI: https://doi.org/10.1261/rna.044032.113
- Primary Citation of Related Structures:
4NL2, 4NL3, 4NOY - PubMed Abstract:
Hfq is a post-transcriptional regulator that binds U- and A-rich regions of sRNAs and their target mRNAs to stimulate their annealing in order to effect translation regulation and, often, to alter their stability. The functional importance of Hfq and its RNA-binding properties are relatively well understood in Gram-negative bacteria, whereas less is known about the RNA-binding properties of this riboregulator in Gram-positive species. Here, we describe the structure of Hfq from the Gram-positive pathogen Listeria monocytogenes in its RNA-free form and in complex with a U6 oligoribonucleotide. As expected, the protein takes the canonical hexameric toroidal shape of all other known Hfq structures. The U6 RNA binds on the "proximal face" in a pocket formed by conserved residues Q9, N42, F43, and K58. Additionally residues G5 and Q6 are involved in protein-nucleic and inter-subunit contacts that promote uracil specificity. Unlike Staphylococcus aureus (Sa) Hfq, Lm Hfq requires magnesium to bind U6 with high affinity. In contrast, the longer oligo-uridine, U16, binds Lm Hfq tightly in the presence or absence of magnesium, thereby suggesting the importance of additional residues on the proximal face and possibly the lateral rim in RNA interaction. Intrinsic tryptophan fluorescence quenching (TFQ) studies reveal, surprisingly, that Lm Hfq can bind (GU)3G and U6 on its proximal and distal faces, indicating a less stringent adenine-nucleotide specificity site on the distal face as compared to the Gram-positive Hfq proteins from Sa and Bacillus subtilis and suggesting as yet uncharacterized RNA-binding modes on both faces.
- Department of Biochemistry, Duke University, Durham, North Carolina 27110, USA.
Organizational Affiliation: