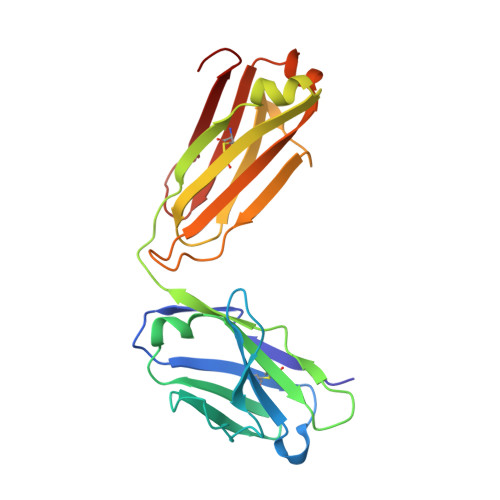
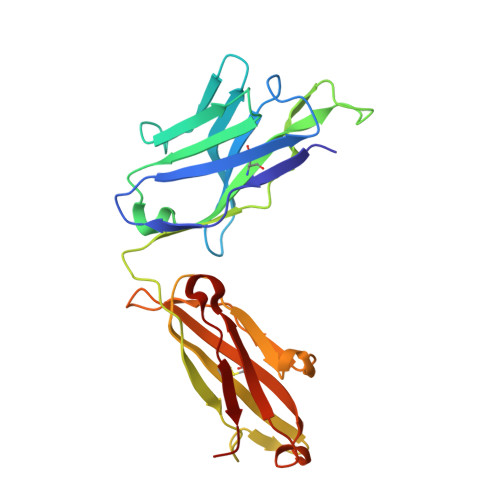
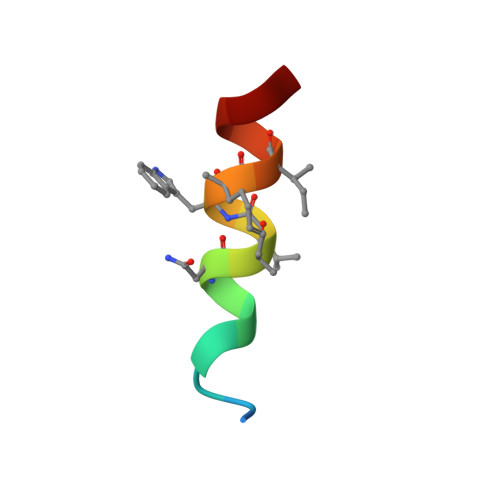
Stapled HIV-1 peptides recapitulate antigenic structures and engage broadly neutralizing antibodies.
Bird, G.H., Irimia, A., Ofek, G., Kwong, P.D., Wilson, I.A., Walensky, L.D.(2014) Nat Struct Mol Biol 21: 1058-1067
- PubMed: 25420104
- DOI: https://doi.org/10.1038/nsmb.2922
- Primary Citation of Related Structures:
4NGH, 4NHC - PubMed Abstract:
Hydrocarbon stapling can restore bioactive α-helical structure to natural peptides, yielding research tools and prototype therapeutics to dissect and target protein interactions. Here we explore the capacity of peptide stapling to generate high-fidelity, protease-resistant mimics of antigenic structures for vaccine development. HIV-1 has been refractory to vaccine technologies thus far, although select human antibodies can broadly neutralize HIV-1 by targeting sequences of the gp41 juxtamembrane fusion apparatus. To develop candidate HIV-1 immunogens, we generated and characterized stabilized α-helices of the membrane-proximal external region (SAH-MPER) of gp41. SAH-MPER peptides were remarkably protease resistant and bound to the broadly neutralizing 4E10 and 10E8 antibodies with high affinity, recapitulating the structure of the MPER epitope when differentially engaged by the two anti-HIV Fabs. Thus, stapled peptides may provide a new opportunity to develop chemically stabilized antigens for vaccination.
- 1] Department of Pediatric Oncology, Dana-Farber Cancer Institute, Boston, Massachusetts, USA. [2] Division of Hematology/Oncology, Boston Children's Hospital, Boston, Massachusetts, USA. [3] Department of Pediatrics, Harvard Medical School, Boston, Massachusetts, USA.
Organizational Affiliation: